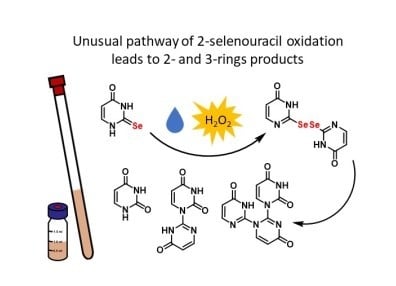
Different Oxidation Pathways of 2-Selenouracil and 2-Thiouracil, Natural Components of Transfer RNA
Abstract
:1. Introduction
2. Results
2.1. General Approach for Analysis of the Course of Oxidation of 2-Selenouracil (1a) and 2-Thiouracil (1b) and Identification of the Reaction Products
2.2. Analysis of the Oxidation Course of Se2Ura (1a)
2.2.1. The Reaction of Se2Ura and H2O2 at a 1:1 Molar Ratio, pH 7.4
2.2.2. The Reaction of Se2Ura and H2O2 at a 1:0.5 Molar Ratio and pH 7.4
2.2.3. The Reaction of Se2Ura and H2O2 at a 1:10 Molar Ratio and pH 7.4
2.3. Analysis of the Oxidation Course of S2Ura (1b)
2.3.1. The Reaction of S2Ura and H2O2 at a 1:1 Molar Ratio and pH 7.4.
2.3.2. The Reaction of S2Ura and H2O2 at a 1:10 Molar Ratio and pH 7.4
3. Discussion
4. Materials and Methods
4.1. Methods and Instrumentation
4.1.1. NMR Spectroscopy
4.1.2. Ultra-Performance Liquid Chromatography Coupled with a High-Resolution Mass Spectrometry and Photodiode Array Detection (UPLC-PDA-ESI(−)-HRMS)
4.1.3. Ultraviolet Spectroscopy Measurements (UV)
4.2. Experimental Section
4.2.1. Materials
4.2.2. Synthesis of 2-Selenouracil
4.2.3. 1H-NMR Analysis of Oxidation Assays of 1a and 1b
4.2.4. UPLC-PDA-ESI (−)-HRMS Analysis of the Oxidation Assays of 1a and 1b
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gilbert, H.F. Molecular and cellular aspects of thiol-disulfide exchange. In Advances in Enzymology and Related Areas of Molecular Biology; Meister, A., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 1990; Volume 63, pp. 69–172. [Google Scholar] [CrossRef]
- Gilbert, H.F. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995, 251, 8–28. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.F. Thiol/disulfide exchange and redox potentials of proteins. In Bioelectrochemistry of Biomacromolecules: Principles and Practice; Lenaz, G., Milazzo, G., Eds.; Birkhäuser Verlag: Basel, Switzerland, 1997; pp. 256–324. [Google Scholar] [CrossRef]
- Hondal, R.J.; Marino, S.M.; Gladyshev, V.N. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid. Redox Signal. 2013, 18, 1675–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, S.; Kumakura, F.; Komatsu, I.; Arai, K.; Onuma, Y.; Hojo, H.; Singh, B.G.; Priyadarsini, K.I.; Iwaoka, M. Antioxidative glutathione peroxidase activity of selenoglutathione. Angew. Chem. 2011, 50, 2125–2128. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigó, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [Green Version]
- Ramming, T.; Hansen, H.G.; Nagata, K.; Ellgaard, L.; Appenzeller-Herzog, C. Gpx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic. Biol. Med. 2014, 70, 106–116. [Google Scholar] [CrossRef]
- Jacob, C.; Giles, G.; Giles, N.M.; Sies, H. Sulfur and selenium: The role of oxidation state in protein structure and function. Angew. Chem. 2003, 42, 4742–4758. [Google Scholar] [CrossRef]
- Wessjohann, L.A.; Schneider, A.; Abbas, M.; Brand, W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol. Chem. 2007, 388, 997–1006. [Google Scholar] [CrossRef]
- Reich, H.J.; Hondal, R.J. Why Nature chose selenium. ACS Chem. Biol. 2016, 11, 821–841. [Google Scholar] [CrossRef]
- Hondal, R.J.; Ruggles, E.L. Differing views of the role of selenium in thioredoxin reductase. Amino Acids 2011, 41, 73–89. [Google Scholar] [CrossRef] [Green Version]
- Maroney, M.; Hondal, R.J. Selenium versus sulfur: Reversibility of chemical reactions and resistance to permanent oxidation in proteins and nucleic acids. Free Radic. Biol. Med. 2018, 127, 228–237. [Google Scholar] [CrossRef]
- Sochacka, E.; Kraszewska, K.; Sochacki, M.; Sobczak, M.; Janicka, M.; Nawrot, B. The 2-thiouridine unit in the RNA strand is desulfured predominantly to 4-pyrimidinone nucleoside under in vitro oxidative stress conditions. Chem. Commun. 2011, 47, 4914–4916. [Google Scholar] [CrossRef] [PubMed]
- Sochacka, E.; Szczepanowski, R.H.; Cypryk, M.; Sobczak, M.; Janicka, M.; Kraszewska, K.; Bartos, P.; Chwialkowska, A.; Nawrot, B. 2-Thiouracil deprived of thiocarbonyl function preferentially base pairs with guanine rather than adenine in RNA and DNA duplexes. Nucleic Acids Res. 2015, 11, 2499–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sochacka, E.; Bartos, P.; Kraszewska, K.; Nawrot, B. Desulfuration of 2-thiouridine with hydrogen peroxide in the physiological pH range 6.6-7.6 is pH-dependent and results in two distinct products. Bioorg. Med. Chem. Lett. 2013, 23, 5803–5805. [Google Scholar] [CrossRef] [PubMed]
- Bartos, P.; Ebenryter-Olbinska, K.; Sochacka, E.; Nawrot, B. The influence of the C5 substituent on the 2-thiouridine desulfuration pathway and the conformational analysis of the resulting 4-pyrimidinone products. Bioorg. Med. Chem. 2015, 23, 5587–5594. [Google Scholar] [CrossRef] [PubMed]
- Sierant, M.; Kulik, K.; Sochacka, E.; Szewczyk, R.; Sobczak, M.; Nawrot, B. Cytochrome C catalyzes the hydrogen peroxide-assisted oxidative desulfuration of 2-thiouridines in transfer RNAs. ChemBiochem 2018, 4, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Payne, N.C.; Geissler, A.; Button, A.; Sasuclark, A.R.; Schroll, A.L.; Ruggles, E.L.; Gladyshev, V.N.; Hondal, R.J. Comparison of the redox chemistry of sulfur- and selenium-containing analogs of uracil. Free Radic. Biol. Med. 2017, 104, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Leszczynska, G.; Cypryk, M.; Gostynski, B.; Sadowska, K.; Herman, P.; Bujacz, G.; Lodyga-Chruscinska, E.; Sochacka, E.; Nawrot, B. C5-Substituted 2-selenouridines ensure efficient base pairing with guanosine; consequences for reading the NNG-3′ synonymous mRNA codons. Int. J. Mol. Sci. 2020, 21, 2882. [Google Scholar] [CrossRef] [Green Version]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Bagiński, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucl. Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.P.; Fabris DAgris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucl. Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [Green Version]
- Rheinboldt, H.; Giesbreoht, E. Unsymmetrische diselenide. Chem. Ber. 1952, 85, 357–368. [Google Scholar] [CrossRef]
- Johnson, T.B.; Edens, C.O. Complex formations between iodine and µ-mercapto-dihydroglyoxalines. J. Am. Chem. Soc. 1942, 64, 2706–2708. [Google Scholar] [CrossRef]
- Marshall, W.; Singh, J. Oxidative inactivation of ethylenethiourea by hypochlorite in alkaline medium. J. Agric. Food Chem. 1977, 25, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- Jackowiak, P.; Nowacka, M.; Strozycki, P.M.; Figlerowicz, M. RNA degradome-its biogenesis and functions. Nucl. Acids Res. 2011, 39, 7361–7370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoniadis, C.D.; Hadjikakou, S.K.; Hadjiliadis, N.; Papakyriakou, A.; Baril, M.; Butler, I.S. Synthesis and structures of Se analogues of the antithyroid drug 6-n-propyl-2-thiouracil and its alkyl derivatives: Formation of dimeric Se-Se compounds and deselenation reactions of charge-transfer adducts of diiodine. Chem. Eur. J. 2006, 12, 6888–6897. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; d’Ischia, M.; Cioffi, F.A. 2-Thiouracil is a selective inhibitor of neuronal nitric oxide synthase antagonising tetrahydrobiopterin-dependent enzyme activation and dimerization. FEBS Lett. 2000, 485, 109–112. [Google Scholar] [CrossRef]
- Reich, H.J.; Hoeger, C.A.; Willis, J.W.W. Organoselenium chemistry. Characterization of reactive intermediates in the selenoxide syn elimination: Selenenic acids and selenolseleninate esters. J. Am. Chem. Soc. 1982, 104, 2936–2937. [Google Scholar] [CrossRef]
- Reich, H.J.; Hoeger, C.A.; Willis, J.W.W. Organoselenium chemistry: A study of intermediates in the fragmentation of aliphatic ketoselenoxides. Characterization of selenoxides, selenenamides and selenolseleninates by 1H-,13C-and 77Se-NMR. Tetrahedron 1985, 41, 4771–4779. [Google Scholar] [CrossRef]
- Ayrey, G.; Barnard, D.; Woodbridge, D.T. The oxidation of organoselenium compounds by ozone. J. Chem. Soc. 1962, 2089–2099. [Google Scholar] [CrossRef]
- Ishii, A.; Takahashi, T.; Nakayama, J. Hydrolysis of an isolable selenoseleninate under acidic and alkaline conditions. Heteroatom Chem. 2001, 12, 198–203. [Google Scholar] [CrossRef]
- Duddeck, H. 77Se NMR spectroscopy and its applications in chemistry. Annu. Reports NMR Spectrosc. 2004, 52, 105–166. [Google Scholar] [CrossRef]
- Mautner, H.G. The synthesis and properties of some selenopurines and selenopyrimidines. J. Am. Chem. Soc. 1956, 78, 5292–5294. [Google Scholar] [CrossRef]















| Compound | UPLC-PDA-ESI(−)-HRMS | UV | 1H NMR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Elemental Composition | Rt [min] | m/z [M-H]− | λmax | H6 [ppm] | H5 [ppm] | JH6-H5 | |||
| Calcd | Found | ||||||||
| 1a | Se2Ura | C4H4N2OSe | 2.00 | 174.9411 | 174.9415 | 306 | 7.61 | 6.21 | 7.7 |
| 1b | S2Ura | C4H4N2OS | 2.14 | 126.9966 | 126.9977 | 271 | 7.59 | 6.11 | 7.5 |
| 2a | Ura-Se-Se-Ura | C8H6N4O2Se2 | 3.61 | 348.8743 | 348.8748 | 275 | 7.88 | 6.26 | 6.6 |
| 2b | Ura-S-S-Ura | C8H6N4O2S2 | 3.35 | 252.9854 | 252.9853 | 228/274 | 7.88 | 6.22 | 6.4 |
| 3a | Ura-Se-Se-Se-Ura | C8H6N4O2Se3 | 4.17 | 428.7908 | 428.7909 | 238 | - | - | - |
| 3b | Ura-S-S-S-Ura | C8H6N4O2S3 | 4.11 | 284.9575 | 284.9576 | - | - | - | - |
| 4a n = 1 | Ura-SeOH | C4H4N2O2Se | 1.99 | 190.9360 | 190.9367 | - | - | - | - |
| 4a n = 2 | Ura-SeO2H | C4H4N2O3Se | 1.21 | 206.9309 | 206.9307 | 230/267 | 8.09 | 6.48 | 6.6 |
| 4b n = 1 | Ura-SOH | C4H4N2O2S | 1.39 | 142.9915 | 142.9921 | 309 | - | - | - |
| 4b n = 2 | Ura-SO2H | C4H4N2O3S | 1.32 | 158.9864 | 158.9865 | 228/275 | 8.07 | 6.49 | 6.8 |
| 4b n = 3 | Ura-SO3H | C4H4N2O4S | 1.11 | 174.9814 | 174.9820 | 232/270 | 8.02 | 6.44 | 6.3 |
| 5 | Ura | C4H4N2O2 | 1.72 | 111.0194 | 111.0190 | 258 | 7.55 | 5.83 | 7.6 |
| 6 | 4HP | C4H4N2O | 1.72 | 95.0245 | 95.0239 | 223 | 8.02 | 6.55 | 7.0 |
| 7a | C8H6N4O2Se | 3.71 | 268.9578 | 268.9584 | 224/275 | - | - | - | |
| 7b | C8H6N4O2S | 3.64 | 221.0133 | 221.0135 | - | - | - | - | |
| 8 | C8H6N4O3 | 1.52 | 205.0362 | 205.0362 | 267 | 8.09 | 6.42 | 8.0 | |
| 7.85 | 5.98 | 6.2 | |||||||
| 9 | C12H8N6O4 | 1.89 | 299.0529 | 299.0534 | 247 | 8.45 | 6.62 | 7.5 | |
| 8.01 | 6.35 | 6.2 | |||||||
| 7.87 | 6.05 | 7.9 | |||||||
| 10 | C4H4N2O4S2 | 1.90 | 206.9534 | 206.9539 | 277 | 7.82 | 5.96 | 7.0 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulik, K.; Sadowska, K.; Wielgus, E.; Pacholczyk-Sienicka, B.; Sochacka, E.; Nawrot, B. Different Oxidation Pathways of 2-Selenouracil and 2-Thiouracil, Natural Components of Transfer RNA. Int. J. Mol. Sci. 2020, 21, 5956. https://doi.org/10.3390/ijms21175956
Kulik K, Sadowska K, Wielgus E, Pacholczyk-Sienicka B, Sochacka E, Nawrot B. Different Oxidation Pathways of 2-Selenouracil and 2-Thiouracil, Natural Components of Transfer RNA. International Journal of Molecular Sciences. 2020; 21(17):5956. https://doi.org/10.3390/ijms21175956
Chicago/Turabian StyleKulik, Katarzyna, Klaudia Sadowska, Ewelina Wielgus, Barbara Pacholczyk-Sienicka, Elzbieta Sochacka, and Barbara Nawrot. 2020. "Different Oxidation Pathways of 2-Selenouracil and 2-Thiouracil, Natural Components of Transfer RNA" International Journal of Molecular Sciences 21, no. 17: 5956. https://doi.org/10.3390/ijms21175956