Adaptive Traits to Improve Durum Wheat Yield in Drought and Crown Rot Environments
Abstract
:1. Introduction
2. Results
2.1. Variation in CR Severity
2.2. Yield and Component Traits in the Presence and Absence of CR
2.3. Association between qSRA-6A and Field Performance
2.4. Association between qCR-6B and Field Performance
2.5. Alignment of QTL Regions Influencing Crown Rot Severity, Water-Use Traits, and Yield
2.6. The Combined Effect of qCR-6B and qSRA-6A on Yield
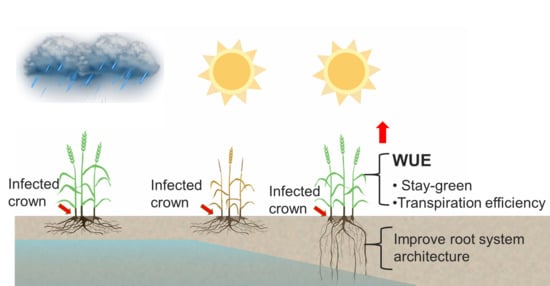
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Establishing a Crown Rot Field Screening Nursery
4.3. Field Experiments
4.3.1. Yield Trials in the Presence of CR
4.3.2. Yield Trials in the Absence of CR
4.4. Phenotyping Root Biomass under Controlled Conditions
4.5. Analysis of Phenotype Data
4.6. Genome-Wide Association Mapping
4.7. Alignment of QTL for Aboveground Traits Across Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Araus, J.L.; Slafer, G.A.; Reynolds, M.P.; Royo, C. Plant breeding and drought in C3 cereals: What should we breed for? Ann. Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.H.; Hewitson, B.; Busuioc, A.; Chen, A.; Gao, X.; Held, R.; Jones, R.K.; Kolli, W.-T.; Kwon, R.; Laprise, V.; et al. Regional Climate Projections. In Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Solomon, S., Qin, D., Manning, M., Chen, Z., Marquis, M., Averyt, K.B., Tignor, M., Miller, H.S., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2007. [Google Scholar]
- Verrell, A.G.; Simpfendorfer, S.; Moore, K.J. Effect of row placement, stubble management and ground engaging tool on crown rot and grain yield in a no-till continuous wheat sequence. Soil Tillage Res. 2017, 165, 16–22. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Qian, L.; Gabur, I.; Obermeier, C.; Hickey, L.T.; Werner, C.R.; Kontowski, S.; Frisch, M.; Friedt, W.; Snowdon, R.J.; et al. Genetic insights into underground responses to Fusarium graminearum infection in wheat. Sci. Rep. 2018, 8, 13153. [Google Scholar] [CrossRef] [PubMed]
- Hollaway, G.J.; Evans, M.L.; Wallwork, H.; Dyson, C.B.; Mckay, A.C. Yield loss in cereals, caused by Fusarium culmorum and f. Pseudograminearum, is related to fungal DNA in soil prior to planting, rainfall, and cereal type. Plant. Dis. 2013, 97, 977–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backhouse, D.; Abubakar, A.A.; Burgess, L.W.; Dennis, J.I.; Hollaway, G.J.; Wildermuth, G.B.; Wallwork, H.; Henry, F.J. Survey of fusarium species associated with crown rot of wheat and barley in eastern Australia. Australas. Plant. Pathol. 2004, 332, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Smiley, R.W.; Patterson, L.M. Pathogenic fungi associated with Fusarium foot rot of winter wheat in the semi-arid Pacific Northwest. Plant. Dis. 1996, 80, 944–949. [Google Scholar] [CrossRef]
- Smiley, R.W.; Gourlie, J.A.; Easley, S.A.; Patterson, L.M.; Whittaker, R.G. Crop damage estimates for crown rot of wheat and barley in the pacific northwest. Plant. Dis. 2005, 89, 595–604. [Google Scholar] [CrossRef] [Green Version]
- Burgess, L.W.; Backhouse, D.; Summerell, B.A. Crown Rot of Wheat. In Fusarium: Paul, E. Nelson Memorial Symposium; Summerell, B.A., Leslie, J.F., Backhouse, D., Bryden, W.L., Burgess, L.W., Eds.; APS Press: St Paul, MN, USA, 2001; pp. 271–294. [Google Scholar]
- Tunali, B.; Nicol, J.; Hodson, D.; Uckun, Z.; Büyük, O.; Erdurmus, D.; Hekimhan, H.; Aktaş, H.; Akbudak, M.; Bağci, A. Root and Crown Rot Fungi Associated with Spring, Facultative, and Winter Wheat in Turkey. Plant. Dis. 2008, 929, 1299–1306. [Google Scholar] [CrossRef]
- Zheng, Z.; Ma, J.; Stiller, J.; Zhao, Q.; Feng, Q.; Choulet, F.; Feuillet, C.; Zheng, Y.; Wei, Y.; Han, B.; et al. Fine mapping of a large-effect QTL conferring Fusarium crown rot resistance on the long arm of chromosome 3B in hexaploid wheat. BMC Genomics 2015, 161, 850. [Google Scholar] [CrossRef] [Green Version]
- Knight, N.L.; Sutherland, M.W. Histopathological assessment of Fusarium pseudograminearum colonization of cereal culms during crown rot infections. Plant. Dis. 2016, 100, 252–259. [Google Scholar] [CrossRef] [Green Version]
- Alahmad, S.; Simpfendorfer, S.; Bentley, A.R.; Hickey, L.T. Crown rot of wheat in Australia: Fusarium pseudograminearum taxonomy, population biology and disease management. Australas. Plant. Pathol. 2018, 47, 285–299. [Google Scholar] [CrossRef]
- Klien, T.A.; Burgeess, L.W.; Ellison, E.W. The incidence and spatial patterns of wheat plants infected by Fusarium graminearum Group 1 and the effect of crown rot on yield. Aust. J. Agric. Res. 1991, 42, 399–407. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant. Path. 2018, 19, 1547–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollaway, G.J.; Exell, G.K. Survey of wheat crops for whiteheads caused by crown rot in victoria, 1997–2009. Australas. Plant. Pathol. 2010, 39, 363–367. [Google Scholar] [CrossRef]
- Chekali, S.; Gargouri, S.; Paulitz, T.; Nicol, J.M.; Rezgui, M.; Nasraoui, B. Effects of fusarium culmorum and water stress on durum wheat in Tunisia. Crop Prot. 2011, 30, 718–725. [Google Scholar] [CrossRef]
- Graham, R. Fusarium crown rot of wheat-impact on plant available soil water usage. In Proceedings of the 17th Australian Agronomy Conference, Hobart, Australia, 20–24 September 2015; pp. 103–105. [Google Scholar]
- Liu, C.J.; Ogbonnaya, F.C. Resistance to fusarium crown rot in wheat and barley: A review. Plant. Breed. 2015, 134, 365–372. [Google Scholar] [CrossRef]
- Liu, X.; Liu, C. Effects of drought-stress on Fusarium crown rot development in Barley. PLoS ONE 2016, 11, e0167304. [Google Scholar] [CrossRef] [Green Version]
- Felton, W.L.; Marcellos, H.; Alston, C.; Martin, R.J.; Backhouse, D.; Burgess, L.W.; Herridge, D.F. Chickpea in wheat-based cropping systems of northern new south wales—II. Influence on biomass, grain yield, and crown rot in the following wheat crop. Aust. J. Agric. Res. 1998, 49, 401–407. [Google Scholar] [CrossRef]
- Wildermuth, G.B.; Mcnamara, R.B.; Quick, J.S. Crown depth and susceptibility to crown rot in wheat. Euphytica 2001, 122, 397–405. [Google Scholar] [CrossRef]
- Simpfendorfer, S.; Kirkegaard, J.A.; Holland, J.; Verrell, A.; Bambach, R.; Moore, K.J. Managing soil and stubble-borne cereal pathogens in the northern grains belt. In Proceedings of the Soil Biology in Agriculture Workshop, Tamworth, Australia, 11–12 August 2004; pp. 112–119. [Google Scholar]
- Verrell, A.; Simpfendorfer, S.; Nash, P.; Moore, K. Can inter-row sowing be used in continuous wheat systems to control crown rot and increase yield? In Proceedings of the 13th Annual Symposium on precision Agriculture in Australasia, Armidale, Australia, 10–11 September 2009. [Google Scholar]
- Simpfendorfer, S.; Brettell, R.; Nicol, J. Inter-row sowing reduces crown rot in winter cereals. In Proceedings of the First International Crown Rot Workshop for wheat improvement (Organising Committee of the 1st International Crown Rot Workshop), Narrabri, Australia, 22–23 October 2012. [Google Scholar]
- Simpfendorfer, S.; Gardner, M. Crown Rot: Be Aware of the Balancing Act or the Fall May Be Harder. GRDC Update Papers 25 Feb. 2013. Available online: http://www.grdc.com.au/Research-and-Development/GRDC-Update-Papers/2013/02/Crown-rot-be-aware-of-the-balancing-act-or-the-fall-may-be-harder (accessed on 22 July 2020).
- Krishnamurthy, L.; Kashiwagi, J.; Upadhyaya, H.D.; Gowda, C.L.L.; Gaur, P.M.; Singh, S.; Purushothaman, R.; Varshney, R.K. Partitioning coefficient—A trait that contributes to drought tolerance in chickpea. Field Crops Res. 2013, 149, 354–365. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, L.; Gaur, P.M.; Basu, P.S.; Chaturvedi, S.K.; Tripathi, S.; Vadez, V.; Rathore, A.; Varshney, R.K.; Gowda, C.L.L. Large genetic variation for heat in the reference collection of chickpea (Cicer arietinum L.) germplasm. Plant. Genet. Resour. 2011, 9, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Christopher, J.; Richard, C.; Chenu, K.; Christopher, M.; Borrell, A.; Hickey, L. Integrating rapid phenotyping and speed breeding to improve stay green and root adaptation of wheat in changing, water-limited, Australian environments. Procedia Environ. Sci. 2015, 29, 175–176. [Google Scholar] [CrossRef] [Green Version]
- Christopher, J.T.; Christopher, M.J.; Borrell, A.K.; Fletcher, S.; Chenu, K. Stay-green traits to improve wheat adaptation in well-watered and water-limited environments. J. Exp. 2016, 67, 5159–5172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gous, P.W.; Hasjim, J.; Franckowiak, J.; Fox, G.P.; Gilbert, R.G. Barley genotype expressing “stay-green”-like characteristics maintains starch quality of the grain during water stress condition. J. Cereal Sci. 2013, 58, 414–419. [Google Scholar] [CrossRef]
- Jordan, D.R.; Hunt, C.H.; Cruickshank, A.W.; Borrell, A.K.; Henzell, R.G. The relationship between the stay-green trait and grain yield in elite sorghum hybrids grown in a range of environments. Crop Sci. 2012, 52, 1153–1161. [Google Scholar] [CrossRef]
- Borrell, A.K.; Van Oosterom, E.J.; Mullet, J.E.; George-Jaeggli, B.; Jordan, D.R.; Klein, P.E.; Hammer, G.L. Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol. 2014, 203, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Hammer, G.L.; Dong, Z.; Mclean, G.; Doherty, A.; Messina, C.; Schussler, J.; Zinselmeier, C.; Paszkiewicz, S.; Cooper, M. Can changes in canopy and/or root system architecture explain historical maize yield trends in the US corn belt? Crop Sci. 2009, 49, 299–312. [Google Scholar] [CrossRef]
- Borrell, A.K.; Mullet, J.E.; George-Jaeggli, B.; Van Oosterom, E.J.; Hammer, G.L.; Klein, P.E.; Jordan, D.R. Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J. Exp. Bot. 2014, 65, 6251–6263. [Google Scholar] [CrossRef]
- Tuberosa, R.; Giuliani, S.; Parry, M.A.J.; Araus, J.L. Improving water use efficiency in Mediterranean agriculture: What limits the adoption of new technologies? Ann. Appl. Biol. 2007, 150, 157–162. [Google Scholar] [CrossRef]
- Maccaferri, M.; El-Feki, W.; Nazemi, G.; Salvi, S.; Cane, M.A.; Colalongo, M.C.; Stefanelli, S.; Tuberosa, R. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J. Exp. 2016, 67, 1161–1178. [Google Scholar] [CrossRef]
- Roselló, M.; Royo, C.; Sanchez-Garcia, M.; Soriano, J.M.J.A. Genetic dissection of the seminal root system architecture in Mediterranean durum wheat landraces by genome-wide association study. Agronomy 2019, 9, 364. [Google Scholar] [CrossRef] [Green Version]
- Alahmad, S.; El Hassouni, K.; Bassi, F.M.; Dinglasan, E.; Youssef, C.; Quarry, G.; Aksoy, A.; Mazzucotelli, E.; Juhasz, A.; Able, J.A.; et al. A major root architecture qtl responding to water limitation in durum wheat. Front. Plant. Sci. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alahmad, S.; Dinglasan, E.; Leung, K.M.; Riaz, A.; Derbal, N.; Voss-Fels, K.P.; Able, J.A.; Bassi, F.M.; Christopher, J.; Hickey, L.T. Speed breeding for multiple quantitative traits in durum wheat. Plant. Methods 2018, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [Green Version]
- Maccaferri, M.; Sanguineti, M.C.; Corneti, S.; Ortega, J.L.; Salem, M.B.; Bort, J.; Deambrogio, E.; Del Moral, L.F.; Demontis, A.; El-Ahmed, A.; et al. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum desf.) across a wide range of water availability. Genetics 2008, 178, 489–511. [Google Scholar] [CrossRef] [Green Version]
- Ghavami, F.; Elias, E.M.; Mamidi, S.; Ansari, O.; Sargolzaei, M.; Adhikari, T.; Mergoum, M.; Kianian, S.F. Mixed model association mapping for fusarium head blight resistance in Tunisian-derived durum wheat populations. G3-Genes Genom Genet. 2011, 1, 209–218. [Google Scholar] [CrossRef]
- Peleg, Z.; Fahima, T.; Korol, A.B.; Abbo, S.; Saranga, Y. Genetic analysis of wheat domestication and evolution under domestication. J. Exp. 2011, 62, 5051–5061. [Google Scholar] [CrossRef] [Green Version]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Giove, S.; Colasuonno, P.; Simeone, R.; Signorile, A.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L.; et al. Relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars. Mol. Breed. 2012, 30, 79–92. [Google Scholar] [CrossRef]
- Prat, N.; Buerstmayr, M.; Steiner, B.; Robert, O.; Buerstmayr, H. Current knowledge on resistance to fusarium head blight in tetraploid wheat. Mol. Breed. 2014, 34, 1689–1699. [Google Scholar] [CrossRef]
- Mengistu, D.K.; Kidane, Y.G.; Catellani, M.; Frascaroli, E.; Fadda, C.; Pè, M.E.; Dell’acqua, M. High-density molecular characterization and association mapping in Ethiopian durum wheat landraces reveal high diversity and potential for wheat breeding. Plant. Biotechnol. J. 2016, 14, 1800–1812. [Google Scholar] [CrossRef] [Green Version]
- Colasuonno, P.; Lozito, M.L.; Marcotuli, I.; Nigro, D.; Giancaspro, A.; Mangini, G.; De Vita, P.; Mastrangelo, A.M.; Pecchioni, N.; Houston, K. The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments. BMC Genomics 2017, 18, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manschadi, A.M.; Christopher, J.; Devoil, P.; Hammer, G.L. The role of root architectural traits in adaptation of wheat to water-limited environments. Funct. Plant. Biol. 2006, 33, 823–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Hassouni, K.; Alahmad, S.; Belkadi, B.; Filali-Maltouf, A.; Hickey, L.; Bassi, F. Root system architecture and its association with yield under different water regimes in durum wheat. Crop Sci. 2018, 58, 2331–2346. [Google Scholar] [CrossRef] [Green Version]
- Robinson, H.; Kelly, A.; Fox, G.; Franckowiak, J.; Borrell, A.; Hickey, L. Root architectural traits and yield: Exploring the relationship in barley breeding trials. Euphytica 2018, 214, 151. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Robinson, H.; Mudge, S.R.; Richard, C.; Newman, S.; Wittkop, B.; Stahl, A.; Friedt, W.; Frisch, M.; Gabur, I.; et al. VERNALIZATION1 modulates root system architecture in wheat and barley. Mol. Plant. 2018, 11, 226–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mace, E.S.; Singh, V.; Van Oosterom, E.J.; Hammer, G.L.; Hunt, C.H.; Jordan, D.R. Qtl for nodal root angle in sorghum (Sorghum bicolor l. Moench) co-locate with qtl for traits associated with drought adaptation. Theor. Appl. Genet. 2012, 124, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Borrell, A.K.; Hammer, G.L.; Henzell, R.G. Does maintaining green leaf area in sorghum improve yield under drought? II. Dry matter production and yield. Crop Sci. 2000, 40, 1037–1048. [Google Scholar] [CrossRef]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 2006, 11, 381. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P. Stay-green in spring wheat can be determined by spectral reflectance measurements (normalized difference vegetation index) independently from phenology. J. Exp. Bot. 2012, 63, 3789–3798. [Google Scholar] [CrossRef] [Green Version]
- Voss-Fels, K.P.; Qian, L.; Parra-Londono, S.; Uptmoor, R.; Frisch, M.; Keeble-Gagnere, G.; Appels, R.; Snowdon, R.J. Linkage drag constrains the roots of modern wheat. Plant. Cell Environ. 2017, 40, 717–725. [Google Scholar] [CrossRef]
- Butler, D.; Cullis, B.R.; Gilmour, A.; Gogel, B. Asreml-R Reference Manual; The State of Queensland, Department of Primary Industries and Fisheries: Brisbane, Australia, 2009. [Google Scholar]
- R Core Team. R Language Definition; R Foundation for Statistical Computing: Vienna, Austria, 2000. [Google Scholar]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; Van Duijn, C.M. Genabel: An r library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stich, B.; Mohring, J.; Piepho, H.P.; Heckenberger, M.; Buckler, E.S.; Melchinger, A.E. Comparison of mixed-model approaches for association mapping. Genetics 2008, 178, 1745–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorrips, R.E. Mapchart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Traits | Crown Rot Present | Crown Rot Absent | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Adjusted Mean | H2 | CV % | Min | Max | Adjusted Mean | H2 | CV % | |
| Yield (GY; t·h−1) | 0.58 | 3.94 | 2.30 | 0.67 | 27.0 | 3.5 | 6.4 | 4.9 | 0.68 | 13.4 |
| Plant height (PH; cm) | 52.9 | 95.8 | 66.7 | 0.45 | 8.7 | 48.0 | 85.5 | 63.4 | 0.79 | 8.9 |
| Time to flowering (DTF; d) | 99.7 | 112.4 | 106.8 | 0.18 | 2.0 | 96.6 | 106.4 | 101.1 | 0.31 | 2.2 |
| Thousand kernel weight (TKW; g) | 28.3 | 48.7 | 37.7 | 0.77 | 11.2 | 32.5 | 53.8 | 41.9 | 0.72 | 11.0 |
| Onset of leaf senescence (OnS; d) | 7.0 | 19.0 | 6.5 | 0.41 | 13.4 | 4.7 | 26.9 | 16.7 | 0.54 | 11.3 |
| Mid-point of leaf senescence (MidS; d) | 5.3 | 26.9 | 16.8 | 0.48 | 26.9 | 15.6 | 31.8 | 25.3 | 0.45 | 13.4 |
| Near completion of leaf senescence (EndS; d) | 19.2 | 47.7 | 30.0 | 0.25 | 16.5 | 29.2 | 43.3 | 36.3 | 0.26 | 8.3 |
| Stay-green integral (SGint) | 16.4 | 29.8 | 23.0 | 0.5 | 11.0 | 20.4 | 30.8 | 24.9 | 0.47 | 9.9 |
| CR severity (CR) | 2.3 | 8.8 | 5.9 | 0.24 | 19.2 | NA | NA | NA | NA | NA |
| 1st reading whiteheads (% WH1) | 1.1 | 65.2 | 21.3 | 0.45 | 65.4 | NA | NA | NA | NA | NA |
| 2nd reading whiteheads (% WH2) | 1.9 | 77.2 | 37.1 | 0.57 | 48.0 | NA | NA | NA | NA | NA |
| Trait a | QTL Name | Marker | Chromosome | cm b | −log10(P) c | Marker Effect d |
|---|---|---|---|---|---|---|
| Plant height | qPH-6B | 995614 | 6B | 2.19 | 3.42 | 5.339 |
| 1008368 | 6B | 2.19 | 3.37 | 5.305 | ||
| EndS | qSG-0.1-6B | 3025934 | 6B | 82.79 | 3.13 | 1.611 |
| SGint | qSG-integral-6B | 3025934 | 6B | 82.79 | 3.01 | 0.750 |
| Yield (t·h−1) | qGY-6B | 1023342 | 6B | 100.50 | 3.26 | −0.215 |
| 2254131 | 6B | 100.50 | 3.12 | −0.203 | ||
| 4992135 | 6B | 100.50 | 3.15 | −0.203 | ||
| WH1 | qWH1-6B | 1023342 | 6B | 100.50 | 3.88 | 7.350 |
| 2254131 | 6B | 100.50 | 3.44 | 6.744 | ||
| 4992135 | 6B | 100.50 | 3.65 | 7.025 | ||
| 1039837 | 6B | 101.26 | 3.41 | 6.800 | ||
| 2256390 | 6B | 101.26 | 3.60 | 6.998 | ||
| WH2 | qWH2-6B | 1023342 | 6B | 100.50 | 3.40 | 8.582 |
| 2254131 | 6B | 100.50 | 3.36 | 8.386 | ||
| 4992135 | 6B | 100.50 | 3.58 | 8.777 | ||
| CR severity | qCR-6B | 4992135 | 6B | 100.13 | 4.14 | −0.590 |
| 1023342 | 6B | 100.50 | 4.20 | −0.598 | ||
| 2254131 | 6B | 100.50 | 3.93 | −0.568 | ||
| 2256390 | 6B | 101.26 | 3.25 | −0.524 | ||
| 1039837 | 6B | 101.26 | 3.10 | −0.508 |
| Abbreviation | Stay-Green Trait | Description |
|---|---|---|
| OnS | Onset of leaf senescence | Number of days from flowering to 90% of greenness |
| MidS | Mid-point of leaf senescence | Number of days from flowering to 50% of greenness |
| EndS | Near completion of leaf senescence | Number of days from flowering to 10% of greenness |
| SGint | Stay-green integral (senescence integral) | Total stay-green parameter referring to the green leaf area duration from flowering to full senescence |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alahmad, S.; Kang, Y.; Dinglasan, E.; Mazzucotelli, E.; Voss-Fels, K.P.; Able, J.A.; Christopher, J.; Bassi, F.M.; Hickey, L.T. Adaptive Traits to Improve Durum Wheat Yield in Drought and Crown Rot Environments. Int. J. Mol. Sci. 2020, 21, 5260. https://doi.org/10.3390/ijms21155260
Alahmad S, Kang Y, Dinglasan E, Mazzucotelli E, Voss-Fels KP, Able JA, Christopher J, Bassi FM, Hickey LT. Adaptive Traits to Improve Durum Wheat Yield in Drought and Crown Rot Environments. International Journal of Molecular Sciences. 2020; 21(15):5260. https://doi.org/10.3390/ijms21155260
Chicago/Turabian StyleAlahmad, Samir, Yichen Kang, Eric Dinglasan, Elisabetta Mazzucotelli, Kai P. Voss-Fels, Jason A. Able, Jack Christopher, Filippo M. Bassi, and Lee T. Hickey. 2020. "Adaptive Traits to Improve Durum Wheat Yield in Drought and Crown Rot Environments" International Journal of Molecular Sciences 21, no. 15: 5260. https://doi.org/10.3390/ijms21155260