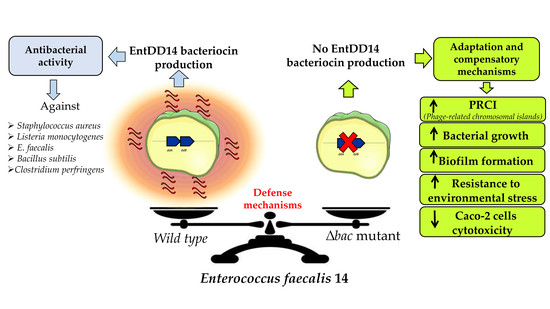
Metabolic Shift of an Isogenic Strain of Enterococcus faecalis 14, Deficient in Its Own Bacteriocin Synthesis, as Revealed by a Transcriptomic Analysis
Abstract
:1. Introduction
2. Results
2.1. Gene Expression Analysis in the E. faecalis 14 Δbac Mutant
2.2. Impact of the Deletion of the Structural Genes Coding for the Bacteriocin on the Bacterial Growth and Ultrastructure
2.3. Absence of EntDD14 and the Biofilm Production Ability of the Δbac Mutant Strain
2.4. Absence of Bacteriocin Reduced the Cytotoxicity of E. faecalis 14
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Antibacterial Activity Assays
4.3. RNA Isolation and Microarrays Analysis
4.4. Transmission Electron Microscopy
4.5. Assessment of Biofilm Formation by Enterococcal Strains on Polystyrene Tissue Culture Plates (TCP)
4.6. CCK-8 Cytotoxicity Assay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; Serrano, L.M.; Griffin, C.; O’Connor, P.M.; Schaad, G.; Bruining, C.; Hill, C.; Ross, R.P.; Meijer, W.C. Inhibitory activity of Lactobacillus plantarum LMG P-26358 against Listeria innocua when used as an adjunct starter in the manufacture of cheese. Microb. Cell Fact. 2011, 10 (Suppl. 1), S7. [Google Scholar] [CrossRef] [Green Version]
- Prokaryotic Antimicrobial Peptides: From Genes to Applications; Drider, D.; Rebuffat, S. (Eds.) Springer: New York, NY, USA, 2011; ISBN 978-1-4419-7691-8. [Google Scholar]
- Besse, A.; Peduzzi, J.; Rebuffat, S.; Carré-Mlouka, A. Antimicrobial peptides and proteins in the face of extremes: Lessons from archaeocins. Biochimie 2015, 118, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Héchard, Y.; Sahl, H.G. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 2002, 84, 545–557. [Google Scholar] [CrossRef]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.S.; Sood, S.K.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef]
- Drissi, F.; Buffet, S.; Raoult, D.; Merhej, V. Common occurrence of antibacterial agents in human intestinal microbiota. Front. Microbiol. 2015, 6, 441. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Drider, D.; Bendali, F.; Naghmouchi, K.; Chikindas, M.L. Bacteriocins: Not only antibacterial agents. Probiotics Antimicrob. Proteins 2016, 8, 177–182. [Google Scholar] [CrossRef]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef]
- Dischinger, J.; Basi Chipalu, S.; Bierbaum, G. Lantibiotics: Promising candidates for future applications in health care. Int. J. Med. Microbiol. 2014, 304, 51–62. [Google Scholar] [CrossRef]
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, potent antimicrobial peptides and the fight against multi drug resistant species: Resistance is futile? Antibiotics (Basel) 2020, 9, 32. [Google Scholar] [CrossRef] [Green Version]
- Drider, D.; Fimland, G.; Héchard, Y.; McMullen, L.M.; Prévost, H. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, M.; Raichaudhuri, A. Bacteriocin in harmony with ABC transporter exhibits antimicrobial activity. EC Microbiol. 2017, 8, 3–10. [Google Scholar]
- Al Atya, A.K.; Drider-Hadiouche, K.; Ravallec, R.; Silvain, A.; Vachee, A.; Drider, D. Probiotic potential of Enterococcus faecalis strains isolated from meconium. Front. Microbiol. 2015, 6, 227. [Google Scholar] [CrossRef]
- Caly, D.L.; Chevalier, M.; Flahaut, C.; Cudennec, B.; Al Atya, A.K.; Chataigné, G.; D’Inca, R.; Auclair, E.; Drider, D. The safe enterocin DD14 is a leaderless two-peptide bacteriocin with anti-Clostridium perfringens activity. Int. J. Antimicrob. Agents 2017, 49, 282–289. [Google Scholar] [CrossRef]
- Ladjouzi, R.; Lucau-Danila, A.; Benachour, A.; Drider, D. A leaderless two-peptide bacteriocin, enterocin DD14, is involved in its own self-immunity: Evidence and insights. Front. Bioeng. Biotechnol. 2020, 8, 644. [Google Scholar] [CrossRef]
- Turgis, M.; Vu, K.D.; Millette, M.; Dupont, C.; Lacroix, M. Influence of environmental factors on bacteriocin production by human isolates of Lactococcus lactis MM19 and Pediococcus acidilactici MM33. Probiotics Antimicrob. Proteins 2016, 8, 53–59. [Google Scholar] [CrossRef]
- Belguesmia, Y.; Leclère, V.; Duban, M.; Auclair, E.; Drider, D. Draft genome sequence of Enterococcus faecalis DD14, a bacteriocinogenic lactic acid bacterium with anti-Clostridium activity. Genome Announc. 2017, 5, e00695-17. [Google Scholar] [CrossRef] [Green Version]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Quintana, V.M.; Torres, N.I.; Wachsman, M.B.; Sinko, P.J.; Castilla, V.; Chikindas, M. Antiherpes simplex virus type 2 activity of the antimicrobial peptide subtilosin. J. Appl. Microbiol. 2014, 117, 1253–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, C.E.; Cruz, M.R.; Garsin, D.A.; Lorenz, M.C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 2017, 114, 4507–4512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.E.M.; Jung, D.Y.-G.; Jin, D.Y.-Y.; Jayabalan, D.R.; Yang, D.S.H.; Suh, P.J.W. Bacteriocins as food preservatives: Challenges and emerging horizons. Crit. Rev. Food Sci. Nutr. 2018, 58, 2743–2767. [Google Scholar] [CrossRef]
- Eijsink, V.G.H.; Axelsson, L.; Diep, D.B.; Håvarstein, L.S.; Holo, H.; Nes, I.F. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie Van Leeuwenhoek 2002, 81, 639–654. [Google Scholar] [CrossRef]
- Birri, D.J.; Brede, D.A.; Forberg, T.; Holo, H.; Nes, I.F. Molecular and genetic characterization of a novel bacteriocin locus in Enterococcus avium isolates from infants. Appl. Environ. Microbiol. 2010, 76, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Matos, R.C.; Lapaque, N.; Rigottier-Gois, L.; Debarbieux, L.; Meylheuc, T.; Gonzalez-Zorn, B.; Repoila, F.; Lopes, M.d.F.; Serror, P. Enterococcus faecalis Prophage dynamics and contributions to pathogenic traits. PLoS Genet. 2013, 9, e1003539. [Google Scholar] [CrossRef] [Green Version]
- Novick, R.P.; Christie, G.E.; Penadés, J.R. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010, 8, 541–551. [Google Scholar] [CrossRef]
- Paulsen, I.T.; Banerjei, L.; Myers, G.S.A.; Nelson, K.E.; Seshadri, R.; Read, T.D.; Fouts, D.E.; Eisen, J.A.; Gill, S.R.; Heidelberg, J.F.; et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2003, 299, 2071–2074. [Google Scholar] [CrossRef] [Green Version]
- Gödeke, J.; Paul, K.; Lassak, J.; Thormann, K.M. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011, 5, 613–626. [Google Scholar] [CrossRef] [Green Version]
- Rice, S.A.; Tan, C.H.; Mikkelsen, P.J.; Kung, V.; Woo, J.; Tay, M.; Hauser, A.; McDougald, D.; Webb, J.S.; Kjelleberg, S. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 2009, 3, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Carrolo, M.; Frias, M.J.; Pinto, F.R.; Melo-Cristino, J.; Ramirez, M. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS ONE 2010, 5, e15678. [Google Scholar] [CrossRef] [Green Version]
- Kristich, C.J.; Li, Y.-H.; Cvitkovitch, D.G.; Dunny, G.M. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 2004, 186, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Guzman, L.-M.; Barondess, J.J.; Beckwith, J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J. Bacteriol. 1992, 174, 7717–7728. [Google Scholar] [CrossRef]
- Sharma, D.; Khan, A.U. Role of cell division protein divIVA in Enterococcus faecalis pathogenesis, biofilm and drug resistance: A future perspective by in silico approaches. Microb. Pathog. 2018, 125, 361–365. [Google Scholar] [CrossRef]
- Afonina, I.; Lim, X.N.; Tan, R.; Kline, K.A. Planktonic interference and biofilm alliance between aggregation substance and endocarditis- and biofilm-associated pili in Enterococcus faecalis. J. Bacteriol. 2018, 200, e00361-18. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-H.; Tang, N.; Aspiras, M.B.; Lau, P.C.Y.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 2002, 184, 2699–2708. [Google Scholar] [CrossRef] [Green Version]
- Ran, S.; Liu, B.; Jiang, W.; Sun, Z.; Liang, J. Transcriptome analysis of Enterococcus faecalis in response to alkaline stress. Front. Microbiol. 2015, 6, 795. [Google Scholar] [CrossRef] [Green Version]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Battán, P.C.; Barnes, A.I.; Albesa, I. Resistance to oxidative stress caused by ceftazidime and piperacillin in a biofilm of Pseudomonas. Luminescence 2004, 19, 265–270. [Google Scholar] [CrossRef]
- Suryaletha, K.; Narendrakumar, L.; John, J.; Radhakrishnan, M.P.; George, S.; Thomas, S. Decoding the proteomic changes involved in the biofilm formation of Enterococcus faecalis SK460 to elucidate potential biofilm determinants. BMC Microbiol. 2019, 19, 146. [Google Scholar] [CrossRef]
- Giard, J.C.; Rince, A.; Capiaux, H.; Auffray, Y.; Hartke, A. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 2000, 182, 4512–4520. [Google Scholar] [CrossRef] [Green Version]
- Carbona, S.L.; Sauvageot, N.; Giard, J.-C.; Benachour, A.; Posteraro, B.; Auffray, Y.; Sanguinetti, M.; Hartke, A. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 2007, 66, 1148–1163. [Google Scholar] [CrossRef]
- Rana, N.F.; Sauvageot, N.; Laplace, J.-M.; Bao, Y.; Nes, I.; Rincé, A.; Posteraro, B.; Sanguinetti, M.; Hartke, A. Redox balance via lactate dehydrogenase is important for multiple stress resistance and virulence in Enterococcus faecalis. Infect. Immun. 2013, 81, 2662–2668. [Google Scholar] [CrossRef] [Green Version]
- Ladjouzi, R.; Bizzini, A.; Lebreton, F.; Sauvageot, N.; Rincé, A.; Benachour, A.; Hartke, A. Analysis of the tolerance of pathogenic enterococci and Staphylococcus aureus to cell wall active antibiotics. J. Antimicrob. Chemother. 2013, 68, 2083–2091. [Google Scholar] [CrossRef]
- Albesa, I.; Becerra, M.C.; Battán, P.C.; Páez, P.L. Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 2004, 317, 605–609. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisithkul, T.; Schroeder, J.W.; Trujillo, E.A.; Yeesin, P.; Stevenson, D.M.; Chaiamarit, T.; Coon, J.J.; Wang, J.D.; Amador-Noguez, D. Metabolic remodeling during biofilm development of bacillus subtilis. mBio 2019, 10, e00623-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, A.; Hardison, R.L.; Wallace, R.M.; Fitch, J.; Heimlich, D.R.; Bryan, M.O.; Dubois, L.; John-Williams, L.S.; Sebra, R.P.; White, P.; et al. Reprioritization of biofilm metabolism is associated with nutrient adaptation and long-term survival of Haemophilus influenzae. npj Biofilms Microbiomes 2019, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Thurlow, L.R.; Thomas, V.C.; Hancock, L.E. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 2009, 191, 6203–6210. [Google Scholar] [CrossRef] [Green Version]
- Trieu-Cuot, P.; Carlier, C.; Poyart-Salmeron, C.; Courvalin, P. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 1991, 102, 99–104. [Google Scholar] [CrossRef]
- Bougherra, F.; Dilmi-Bouras, A.; Balti, R.; Przybylski, R.; Adoui, F.; Elhameur, H.; Chevalier, M.; Flahaut, C.; Dhulster, P.; Naima, N. Antibacterial activity of new peptide from bovine casein hydrolyzed by a serine metalloprotease of Lactococcus lactis subsp lactis BR16. J. Funct. Foods 2017, 32, 112–122. [Google Scholar] [CrossRef]
- Ait Ouali, F.; Al Kassaa, I.; Cudennec, B.; Abdallah, M.; Bendali, F.; Sadoun, D.; Chihib, N.-E.; Drider, D. Identification of lactobacilli with inhibitory effect on biofilm formation by pathogenic bacteria on stainless steel surfaces. Int. J. Food Microbiol. 2014, 191, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.; Robineleon, S.; Appay, M.D.; Kedinger, M.; Triadou, N.; Dussaulx, E.; Lacroix, B.; Simonassmann, P.; Haffen, K.; Fogh, J.; et al. Enterocyte-like differentiation and polarization of the human-colon carcinoma cell-line Caco-2 in culture. Biol. Cell 1983, 47, 323–330. [Google Scholar]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ladjouzi, R.; Lucau-Danila, A.; Drider, D. Metabolic Shift of an Isogenic Strain of Enterococcus faecalis 14, Deficient in Its Own Bacteriocin Synthesis, as Revealed by a Transcriptomic Analysis. Int. J. Mol. Sci. 2020, 21, 4653. https://doi.org/10.3390/ijms21134653
Ladjouzi R, Lucau-Danila A, Drider D. Metabolic Shift of an Isogenic Strain of Enterococcus faecalis 14, Deficient in Its Own Bacteriocin Synthesis, as Revealed by a Transcriptomic Analysis. International Journal of Molecular Sciences. 2020; 21(13):4653. https://doi.org/10.3390/ijms21134653
Chicago/Turabian StyleLadjouzi, Rabia, Anca Lucau-Danila, and Djamel Drider. 2020. "Metabolic Shift of an Isogenic Strain of Enterococcus faecalis 14, Deficient in Its Own Bacteriocin Synthesis, as Revealed by a Transcriptomic Analysis" International Journal of Molecular Sciences 21, no. 13: 4653. https://doi.org/10.3390/ijms21134653