Overexpression of a S-Adenosylmethionine Decarboxylase from Sugar Beet M14 Increased Araidopsis Salt Tolerance
Abstract
:1. Introduction
2. Results
2.1. Isolation of BvM14- SAMDC and Sequence Analysis
2.2. Prokaryotic Expression and Enzymatic Activity Assay of Recombinant BvM14-SAMDC
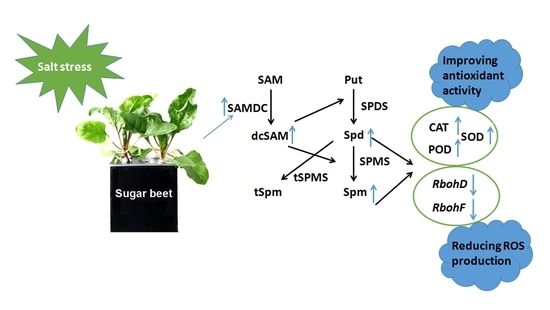
2.3. Expression Patterns of BvM14-SAMDC under Salt Stress
2.4. Overexpression of BvM14-SAMDC Increased Salt Stress Tolerance in Arabidopsis
2.5. Overexpression of BvM14-SAMDC Enhanced Antioxidative Activities in Arabidopsis
2.6. Overexpression of BvM14-SAMDC Enhanced PA Levels and PAO Activity in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Cloning the Ful- Length cDNA of BvM14-SAMDC Gene
4.3. Real-time Quantitative PCR
4.4. Heterologous Expression of BvM14-SAMDC in E.coli
4.5. Arabidopsis Thaliana Mutant Screening
4.6. Generation of 35S:BvM14-SAMDC Construct and Transformation of Arabidopsis
4.7. Determination of Physiological Indicators
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Yusuf, M.A.; Nigam, M.; Kumar, M. An update on genetic modification of chickpea for increased yield and stress tolerance. Mol. Biotechnol. 2018, 60, 651–663. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Shah, N.; Anwar, S.; Xu, J.; Hou, Z.; Salah, A.; Khan, S.; Gong, J.; Shang, Z.; Qian, L.; Zhang, C. The response of transgenic Brassica species to salt stress: A review. Biotechnol. Lett. 2018, 40, 1159–1165. [Google Scholar] [CrossRef]
- Wu, B.; Munkhtuya, Y.; Li, J.; Hu, Y.; Zhang, Q.; Zhang, Z. Comparative transcriptional profiling and physiological responses of two contrasting oat genotypes under salt stress. Sci. Rep. 2018, 8, 16248. [Google Scholar] [CrossRef]
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381. [Google Scholar] [CrossRef]
- Liu, J.H.; Wang, W.; Wu, H.; Gong, X.; Moriguchi, T. Polyamines function in stress tolerance: From synthesis to regulation. Front. Plant Sci. 2015, 6, 827. [Google Scholar] [CrossRef]
- Guo, J.; Wang, S.; Yu, X.; Dong, R.; Li, Y.; Mei, X.; Shen, Y. Polyamines regulate strawberry fruit ripening by abscisic acid, auxin, and ethylene. Plant Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef]
- Mehta, R.A.; Cassol, T.; Li, N.; Ali, N.; Handa, A.K.; Mattoo, A.K. Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality, and vine life. Nat. Biotechnol. 2002, 20, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Angelini, R.; Cona, A.; Federico, R.; Fincato, P.; Tavladoraki, P.; Tisi, A. Plant amine oxidases “on the move”: An update. Plant Physiol. Biochem. 2010, 48, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Roje, S. S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698. [Google Scholar] [CrossRef]
- Romero, F.M.; Maiale, S.J.; Rossi, F.R.; Marina, M.; Ruíz, O.A.; Gárriz, A. Polyamine metabolism responses to biotic and abiotic stress. Methods Mol. Biol. 2018, 1694, 37–49. [Google Scholar] [CrossRef]
- Shu, S.; Yuan, Y.; Chen, J.; Sun, J.; Zhang, W.; Tang, Y.; Zhong, M.; Guo, S. The role of putrescine in the regulation of proteins and fatty acids of thylakoid membranes under salt stress. Sci. Rep. 2015, 5, 14390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Kim, S.H.; Palaniyandi, S.A.; Yang, S.H.; Suh, J.W. Expression of potato S-adenosyl-L-methionine synthase (SbSAMS) gene altered developmental characteristics and stress responses in transgenic Arabidopsis plants. Plant Physiol. Biochem. 2015, 87, 84–91. [Google Scholar] [CrossRef]
- Tanou, G.; Ziogas, V.; Belghazi, M.; Christou, A.; Filippou, P.; Job, D.; Fotopoulos, V.; Molassiotis, A. Polyamines reprogram oxidative and nitrosative status and the proteome of citrus plants exposed to salinity stress. Plant Cell Environ. 2014, 37, 864–885. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Li, X.; VandenLangenberg, K.M.; Wen, D.; Sun, S.; Wei, M.; Li, Y.; Yang, F.; Shi, Q.; Wang, X. Overexpression of S-adenosyl-L-methionine synthetase increased tomato tolerance to alkalistress through polyamine metabolism. Plant Biotechnol. J. 2014, 12, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, Y.; Gu, D.; Nan, J.; Chen, S.; Li, H. Overexpression of S-Adenosyl-l-Methionine Synthetase 2 from sugar beet M14 increased Arabidopsis tolerance to salt and oxidative stress. Int. J. Mol. Sci. 2017, 18, 847. [Google Scholar] [CrossRef] [PubMed]
- Mellidou, I.; Moschou, P.N.; Ioannidis, N.E.; Pankou, C.; Gėmes, K.; Valassakis, C.; Andronis, E.A.; Beris, D.; Haralampidis, K.; Roussis, A.; et al. Silencing S-Adenosyl-L-Methionine Decarboxylase (SAMDC) in Nicotiana tabacum points at a polyamine-dependent trade-off between growth and tolerance responses. Front. Plant Sci. 2016, 7, 379. [Google Scholar] [CrossRef]
- Majumdar, R.; Lebar, M.; Mack, B.; Minocha, R.; Minocha, S.; Carter-Wientjes, C.; Sickler, C.; Rajasekaran, K.; Cary, J.W. The Aspergillus flavus spermidine synthase (spds) gene, is required for normal development, aflatoxin productionand pathogenesis during infection of maize kernels. Front. Plant Sci. 2018, 9, 317. [Google Scholar] [CrossRef]
- Mo, H.J.; Sun, Y.X.; Zhu, X.L.; Wang, X.F.; Zhang, Y.; Yang, J.; Yan, G.J.; Ma, Z.Y. Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid-and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 2016, 243, 1023–1039. [Google Scholar] [CrossRef]
- Ge, Y.; He, G.; Wang, Z.; Guo, D.; Qin, R.; Li, R. GISH and BAC-FISH study of apomicitic Beta M14. Sci. China Ser. C Life Sci. 2007, 37, 209–216. [Google Scholar]
- Guo, D.; Kang, C.; Liu, L.; Li, Y. Study of apomixis in theallotriploid beet (VVC). Agric. Sci. China 1999, 32, 1–5. [Google Scholar]
- Yang, L.; Ma, C.; Wang, L.; Chen, S.; Li, H. Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14. J. Plant Physiol. 2012, 169, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, Y.; Zhu, N.; Koh, J.; Ma, C.; Pan, Y.; Yu, B.; Chen, S.; Li, H. Proteomic analysis of salt tolerance in sugar beet monosomic addition line M14. J. Proteome Res. 2013, 12, 4931–4950. [Google Scholar] [CrossRef]
- Yin, J.; Jia, J.; Lian, Z.; Hu, Y.; Guo, J.; Huo, H.; Zhu, Y.; Gong, H. Silicon enhances the salt tolerance of cucumber through increasing polyamine accumulation and decreasing oxidative damage. Ecotoxicol. Environ. Saf. 2019, 169, 8–17. [Google Scholar] [CrossRef]
- Murray Stewart, T.; Dunston, T.T.; Woster, P.M.; Casero, R.A., Jr. Polyamine catabolism and oxidative damage. J. Biol. Chem. 2018, 293, 18736–18745. [Google Scholar] [CrossRef] [PubMed]
- Wi, S.J.; Kim, S.J.; Kim, W.T.; Park, K.Y. Constitutive S-adenosylmethionine decarboxylase gene expression increases drought tolerance through inhibition of reactive oxygen species accumulation in Arabidopsis. Planta 2014, 239, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Marco, F.; Busó, E.; Carrasco, P. Overexpression of SAMDC1 gene in Arabidopsis increases expression of defense related genes as well as resistance to Pseudomonas syringae and Hyaloperonospora arabidopsidis. Front. Plant Sci. 2014, 5, 115. [Google Scholar] [CrossRef]
- Ibrahim, W.; Qiu, C.W.; Zhang, C.; Cao, F.; Shuijin, Z.; Wu, F. Comparative physiological analysis in the tolerance to salinity and drought individual and combination in two cotton genotypes with contrasting salt tolerance. Physiol. Plant. 2019, 165, 155–168. [Google Scholar] [CrossRef]
- Groppa, M.D.; Benavides, M.P. Polyamines and abiotic stress: Recent advances. Amino Acids 2008, 34, 35–45. [Google Scholar] [CrossRef]
- Cona, A.; Rea, G.; Botta, M.; Corelli, F.; Federico, R.; Angelini, R. Flavin-containing polyamine oxidase is a hydrogen peroxide source in the oxidative response to the protein phosphatase inhibitor cantharidin in Zea mays L. J. Exp. Bot. 2006, 57, 2277–2289. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, E.; Martínez, J.P.; Benahmed, H.; Fauconnier, M.L.; Lutts, S.; Quinet, M. Salicylic acid differently impacts ethylene and polyamine synthesis in the glycophyte Solanum lycopersicum and the wild-related halophyte Solanum chilense exposed to mild salt stress. Physiol. Plant. 2016, 158, 152–167. [Google Scholar] [CrossRef]
- Botella, M.A.; Xu, Y.; Prabha, T.N.; Zhao, Y.; Narasimhan, M.L.; Wilson, K.A.; Nielsen, S.S.; Bressan, R.A.; Hasegawa, P.M. Differential expression of soybean cysteine proteinase inhibitor genes during development and in response to wounding and methyl jasmonate. Plant Physiol. 1996, 112, 1201–1210. [Google Scholar] [CrossRef] [Green Version]
- Hazarika, P.; Rajam, M.V. Biotic and abiotic stress tolerance in transgenic tomatoes by constitutive expression of S-adenosylmethionine decarboxylase gene. Physiol. Mol. Biol. Plants 2011, 17, 115–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wi, S.J.; Kim, W.T.; Park, K.Y. Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep. 2006, 25, 1111–1121. [Google Scholar] [CrossRef]
- Waie, B.; Rajam, M.V. Effect of increased polyamine biosynthesis on stress responses in transgenic tobacco by introduction of human S-adenosylmethionine gene. Plant Sci. 2003, 164, 727–734. [Google Scholar] [CrossRef]
- Roy, M.; Wu, R. Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride-stress tolerance. Plant Sci. 2002, 163, 987–992. [Google Scholar] [CrossRef]
- Yang, L.; Ye, C.; Zhao, Y.; Cheng, X.; Wang, Y.; Jiang, Y.Q.; Yang, B. An oilseed rape WRKY-type transcription factor regulates ROS accumulation and leaf senescence in Nicotiana benthamiana and Arabidopsis through modulating transcription of RbohD and RbohF. Planta 2018, 247, 1323–1338. [Google Scholar] [CrossRef]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 2016, 67, 1663–1676. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhan, Y.; Wu, C.; Gong, S.; Zhu, N.; Chen, S.; Li, H. Cloning of a cystatin gene from sugar beet M14 that can enhance plant salt tolerance. Plant Sci. 2012, 191–192, 93–99. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Aebi, H. CAT in vitro. Meth. Enzymol. 1984, 105, 121–126. [Google Scholar] [CrossRef]
- Wang, Y.; Stevanato, P.; Yu, L.; Zhao, H.; Sun, X.; Sun, F.; Li, J.; Geng, G. The physiological and metabolic changes in sugar beet seedlings under different levels of salt stress. J. Plant Res. 2017, 130, 1079–1093. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, M.; Wang, K.; Wang, L.; Chen, S.; Li, H.; Ma, C.; Wang, Y. Overexpression of a S-Adenosylmethionine Decarboxylase from Sugar Beet M14 Increased Araidopsis Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 1990. https://doi.org/10.3390/ijms20081990
Ji M, Wang K, Wang L, Chen S, Li H, Ma C, Wang Y. Overexpression of a S-Adenosylmethionine Decarboxylase from Sugar Beet M14 Increased Araidopsis Salt Tolerance. International Journal of Molecular Sciences. 2019; 20(8):1990. https://doi.org/10.3390/ijms20081990
Chicago/Turabian StyleJi, Meichao, Kun Wang, Lin Wang, Sixue Chen, Haiying Li, Chunquan Ma, and Yuguang Wang. 2019. "Overexpression of a S-Adenosylmethionine Decarboxylase from Sugar Beet M14 Increased Araidopsis Salt Tolerance" International Journal of Molecular Sciences 20, no. 8: 1990. https://doi.org/10.3390/ijms20081990