Serine/Threonine Protein Kinase STK16
Abstract
:1. Introduction
2. Distribution and Characterization of STK16
2.1. Distribution
2.2. Molecular Characterization of STK16
3. Posttranslational Modification of STK16
3.1. STK16 Undergoes Specific Fatty Acylation Modification
3.2. STK16 is a Constitutively Active Ser/Thr Protein Kinase
3.3. The Relationship between Fatty Acylation and Phosphorylation of STK16
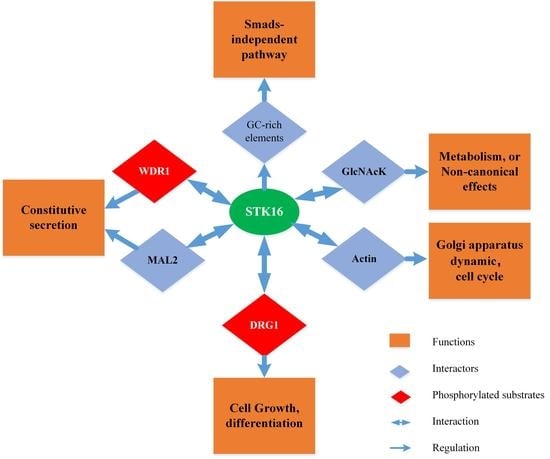
4. Cellular Functions of STK16
4.1. STK16 Participates in the TGF- β Signaling Pathway
4.2. STK16 and GlcNAcK (N-Acetylglucosamine Kinase)
4.3. STK16 and MAL2/WDR1 (Myelin and Lymphocyte Protein 2/ WD Repeat Containing Protein-1)
4.4. STK16 and DRG1 (Developmentally Regulated GTP Binding Protein 1)
4.5. STK16 and Actin
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Wilson, L.J.; Linley, A.; Hammond, D.E.; Hood, F.E.; Coulson, J.M.; Macewan, D.J.; Ross, S.J.; Slupsky, J.R.; Smith, P.D.; Eyers, P.A. New Perspectives, Opportunities, and Challenges in Exploring the Human Protein Kinome. Cancer Res. 2018, 78, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Gulluni, F.; De Santis, M.C.; Margaria, J.P.; Martini, M.; Hirsch, E. Class II PI3K Functions in Cell Biology and Disease. Trends Cell Biol. 2019, 29, 339–359. [Google Scholar] [CrossRef]
- Fu, J.; Bian, M.; Jiang, Q.; Zhang, C. Roles of Aurora Kinases in Mitosis and Tumorigenesis. Mol. Cancer Res. 2007, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Sui, X.; Kong, N.; Ye, L.; Han, W.; Zhou, J.; Zhang, Q.; He, C.; Pan, H. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014, 344, 174–179. [Google Scholar] [CrossRef] [PubMed]
- García, Z.; Kumar, A.; Marques, M.; Cortes, I.; Carrera, A.C. Phosphoinositide 3-kinase controls early and late events in mammalian cell division. Embo J. 2006, 25, 655–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanks, S.K.; Quinn, A.M. Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991, 200, 38–62. [Google Scholar] [PubMed]
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 1988, 241, 42–52. [Google Scholar] [CrossRef]
- AbdElsalam, K.A. Bioinformatic tools and guideline for PCR primer design. Afr. J. Biotechnol. 2003, 2, 91–95. [Google Scholar]
- Bachman, J. Reverse-transcription PCR (RT-PCR). Methods Enzymol. 2013, 530, 67–74. [Google Scholar] [PubMed]
- Ligos, J.M.; Gerwin, N.; Fernández, P.; Gutierrez-Ramos, J.C.; Bernad, A. Cloning, Expression Analysis, and Functional Characterization of PKL12, a Member of a New Subfamily of ser/thr Kinases. Biochem. Biophys. Res. Commun. 1998, 249, 380–384. [Google Scholar] [CrossRef]
- Stairs, D.B.; Gardner, H.P.; Ha, S.I.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Chodosh, L.A. Cloning and characterization of Krct, a member of a novel subfamily of serine/threonine kinases. Hum. Mol. Genet. 1998, 7, 2157–2166. [Google Scholar] [CrossRef] [Green Version]
- Kurioka, K.; Nakagawa, K.; Denda, K.; Miyazawa, K.; Kitamura, N. Molecular cloning and characterization of a novel protein serine/threonine kinase highly expressed in mouse embryo 1. Biochim. Biophys. Acta 1998, 1443, 275–284. [Google Scholar] [CrossRef]
- Berson, A.E.; Young, C.; Morrison, S.L.; Fujii, G.H.; Sheung, J.; Wu, B.; Bolen, J.B.; Burkhardt, A.L. Identification and characterization of a myristylated and palmitylated serine/threonine protein kinase. Biochem. Biophys. Res. Commun. 1999, 259, 533–538. [Google Scholar] [CrossRef]
- Ohta, S.; Takeuchi, M.; Deguchi, M.; Tsuji, T.; Gahara, Y.; Nagata, K. A novel transcriptional factor with Ser/Thr kinase activity involved in the transforming growth factor (TGF)-beta signalling pathway. Biochem. J. 2000, 350, 395–404. [Google Scholar] [CrossRef]
- Sorrell, F.; Szklarz, M.; Abdulazeez, K.; Elkins, J.; Knapp, S. Family-wide Structural Analysis of Human Numb-Associated Protein Kinases. Structure 2016, 24, 401–411. [Google Scholar] [CrossRef] [Green Version]
- Eswaran, J.; Bernad, A.; Ligos, J.M.; Guinea, B.; Debreczeni, J.É.; Sobott, F.; Parker, S.; Najmanovich, R.; Turk, B.E.; Knapp, S. Structure of the Human Protein Kinase MPSK1 Reveals an Atypical Activation Loop Architecture. Structure 2008, 16, 115–124. [Google Scholar] [CrossRef]
- Stairs, D.B.; Notarfrancesco, K.L.; Chodosh, L.A. The serine/threonine kinase, Krct, affects endbud morphogenesis during murine mammary gland development. Transgenic Res. 2005, 14, 919–940. [Google Scholar] [CrossRef]
- Guinea, B.; Ligos, J.M.; Lera, T.L.D.; Martín-Caballero, J.; Flores, J.; Peña, M.G.D.L.; García-Castro, J.; Bernada, A. Nucleocytoplasmic shuttling of STK16 (PKL12), a Golgi-resident serine/threonine kinase involved in VEGF expression regulation. Exp. Cell Res. 2006, 312, 135–144. [Google Scholar] [CrossRef]
- Ligos, J.M.; de Lera, T.L.; Hinderlich, S.; Guinea, B.; Sánchez, L.; Roca, R.; Valencia, A.; Bernad, A. Functional interaction between the Ser/Thr kinase PKL12 and N-acetylglucosamine kinase, a prominent enzyme implicated in the salvage pathway for GlcNAc recycling. J. Biol. Chem. 2002, 277, 6333–6343. [Google Scholar] [CrossRef]
- Liu, J.; Yang, X.; Li, B.; Wang, J.; Wang, W.; Liu, J.; Liu, Q.; Zhang, X. STK16 regulates actin dynamics to control Golgi organization and cell cycle. Sci. Rep. 2017, 7, 44607. [Google Scholar] [CrossRef] [Green Version]
- Florante, R.; Rosa, M.; Surya, C.; Tattika, S.; Lisa, O.; Teli, H.; Maribeth, S.; Editte, G. A genome-wide immunodetection screen in S. cerevisiae uncovers novel genes involved in lysosomal vacuole function and morphology. PLoS ONE 2011, 6, e23696. [Google Scholar]
- Hanks, S.K.; Hunter, T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. Faseb J. 1995, 9, 576–596. [Google Scholar] [CrossRef]
- Chien, C.T.; Wang, S.; Rothenberg, M.; Jan, L.Y.; Jan, Y.N. Numb-associated kinase interacts with the phosphotyrosine binding domain of Numb and antagonizes the function of Numb in vivo. Mol. Cell. Biol. 1998, 18, 598–607. [Google Scholar] [CrossRef]
- Rhyu, M.S.; Jan, L.Y.; Jan, Y.N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 1994, 76, 477–491. [Google Scholar] [CrossRef]
- Uemura, T.; Shepherd, S.; Ackerman, L.; Jan, L.Y.; Jan, Y.N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 1989, 58, 349–360. [Google Scholar] [CrossRef]
- Conner, S.D.; Schmid, S.L. Identification of an adaptor-associated kinase, AAK1, as a regulator of clathrin-mediated endocytosis. J. Cell Biol. 2002, 156, 921–929. [Google Scholar] [CrossRef] [Green Version]
- Conner, S.D.; Schröter, T.; Schmid, S.L. AAK1-mediated micro2 phosphorylation is stimulated by assembled clathrin. Traffic 2010, 4, 885–890. [Google Scholar] [CrossRef]
- Neetu, G.R.; Sara, O.; Vannary, M.Y.; Sara, H.; Julien, M.; Jean-Christophe, O.M.; Alain, I.L. The adaptor-associated kinase 1, AAK1, is a positive regulator of the Notch pathway. J. Biol. Chem. 2011, 286, 18720–18730. [Google Scholar]
- Sorensen, E.B.; Conner, S.D. AAK1 regulates Numb function at an early step in clathrin-mediated endocytosis. Traffic 2010, 9, 1791–1800. [Google Scholar] [CrossRef]
- Krieger, J.R.; Paul, T.; Gajadhar, A.S.; Abhijit, G.; Moran, M.F.; C Jane, M. Identification and selected reaction monitoring (SRM) quantification of endocytosis factors associated with Numb. Mol. Cell. Proteom. 2013, 12, 499–514. [Google Scholar] [CrossRef]
- Lee, D.-W.; Zhao, X.; Zhang, F.; Eisenberg, E.; Greene, L.E. Depletion of GAK/auxilin 2 inhibits receptor-mediated endocytosis and recruitment of both clathrin and clathrin adaptors. J. Cell Sci. 2005, 118, 4311–4321. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Engqvist-Goldstein, A.; Carreno, S.; Owen, D.; Smythe, E.; Drubin, D. Multiple roles for cyclin G-associated kinase in clathrin-mediated sorting events. Traffic 2010, 6, 1103–1113. [Google Scholar] [CrossRef]
- Kostich, W.; Hamman, B.D.; Li, Y.W.; Naidu, S.; Dandapani, K.; Feng, J.; Easton, A.; Bourin, C.; Baker, K.; Allen, J. Inhibition of AAK1 Kinase as a Novel Therapeutic Approach to Treat Neuropathic Pain. J. Pharmacol. Exp. Ther. 2016, 358, 371–386. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Magid, A.F. Inhibitors of Adaptor-Associated Kinase 1 (AAK1) May Treat Neuropathic Pain, Schizophrenia, Parkinson’s Disease, and Other Disorders. ACS Med. Chem. Lett. 2017, 8, 595–597. [Google Scholar] [CrossRef]
- Bekerman, E.; Neveu, G.; Shulla, A.; Brannan, J.; Pu, S.Y.; Wang, S.; Xiao, F.; Barouch-Bentov, R.; Bakken, R.R.; Mateo, R. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Investig. 2017, 127, 1338–1352. [Google Scholar] [CrossRef]
- Sona, K.; Lei, C.; Elena, B.; Gregory, N.; Rina, B.B.; Apirat, C.; Christina, H.; Michal, Á.; Steven, D.J.; Stefan, K. Selective Inhibitors of Cyclin G Associated Kinase (GAK) as Anti-Hepatitis C Agents. J. Med. Chem. 2015, 58, 3393–3410. [Google Scholar]
- Pu, S.Y.; Wouters, R.; Schor, S.; Rozenski, J.; Barouch-Bentov, R.; Prugar, L.I.; Obrien, C.M.; Brannan, J.M.; Dye, J.M.; Herdewijn, P. Optimization of isothiazolo[4,3-b]pyridine-based inhibitors of cyclin G associated kinase (GAK) with broad-spectrum antiviral activity. J. Med. Chem. 2018, 61, 6178–6192. [Google Scholar] [CrossRef]
- Zhou, H.; Min, X.; Qian, H.; Gates, A.T.; Zhang, X.D.; Castle, J.C.; Stec, E.; Ferrer, M.; Strulovici, B.; Hazuda, D.J. Genome-Scale RNAi Screen for Host Factors Required for HIV Replication. Cell Host Microbe 2008, 4, 495–504. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Wang, J.; Yang, X.; Li, B.; Wu, H.; Qi, S.; Chen, C.; Liu, X.; Yu, K.; Wang, W. Discovery of a Highly Selective STK16 Kinase Inhibitor. ACS Chem. Biol. 2016, 11, 1537–1543. [Google Scholar] [CrossRef]
- Anderson, A.M.; Ragan, M.A. Palmitoylation: A protein S-acylation with implications for breast cancer. NPJ Breast Cancer 2016, 2, 16028. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, B.; Dimmock, J.R.; Sharma, R.K. N-myristoyltransferase in the leukocytic development processes. Cell Tissue Res. 2011, 345, 203–211. [Google Scholar] [CrossRef] [Green Version]
- Udenwobele, D.I.; Su, R.-C.; Good, S.V.; Ball, T.B.; Varma Shrivastav, S.; Shrivastav, A. Myristoylation: An important protein modification in the immune response. Front. Immunol. 2017, 8, 751. [Google Scholar] [CrossRef]
- Resh, M.D. Covalent lipid modifications of proteins. Curr. Biol. CB 2013, 23, R431–R435. [Google Scholar] [CrossRef]
- Resh, M.D. Fatty Acylation of Proteins: The Long and the Short of it. Prog. Lipid Res. 2016, 63, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Zhang, X.; Chen, X.; Aramsangtienchai, P.; Tong, Z.; Lin, H. Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies. Chem. Rev. 2018, 118, 919–988. [Google Scholar] [CrossRef]
- Aicart-Ramos, C.; Valero, R.A.; Rodriguez-Crespo, I. Protein palmitoylation and subcellular trafficking. BBA-Biomembr. 2011, 1808, 2981–2994. [Google Scholar] [CrossRef] [Green Version]
- Ryo, M.; Miki, M.; Takefumi, U.; Satoshi, W.; Eiji, M.; Naoyuki, T.; Michiyuki, M.; Tomohiko, T. Palmitoylated Ras proteins traffic through recycling endosomes to the plasma membrane during exocytosis. Autophagy 2011, 191, 23–29. [Google Scholar]
- Stoffel, R.H.; Randall, R.R.; Premont, R.T.; Lefkowitz, R.J.; Inglese, J. Palmitoylation of G protein-coupled receptor kinase, GRK6. Lipid modification diversity in the GRK family. J. Biol. Chem. 1994, 269, 27791–27794. [Google Scholar]
- Konitsiotis, A.D.; Roßmannek, L.; Stanoev, A.; Schmick, M.; Pih, B. Spatial cycles mediated by UNC119 solubilisation maintain Src family kinases plasma membrane localisation. Nat. Commun. 2017, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Shenoy-Scaria, A.M.; Dietzen, D.J.; Kwong, J.; Link, D.C.; Lublin, D.M. Cysteine3 of Src family protein tyrosine kinase determines palmitoylation and localization in caveolae. J. Cell Biol. 1994, 126, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Burgers, P.P.; Yuliang, M.; Luigi, M.; Mason, M.; Van Der Heyden, M.A.G.; Mark, E.; Arjen, S.; Taylor, S.S.; Heck, A.J.R. A small novel A-kinase anchoring protein (AKAP) that localizes specifically protein kinase A-regulatory subunit I (PKA-RI) to the plasma membrane. J. Biol. Chem. 2012, 287, 43789–43797. [Google Scholar] [CrossRef]
- Martin, M.L.; Busconi, L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) ismediated by myristoylation and palmitoylation. Plant J. 2010, 24, 429–435. [Google Scholar]
- Manandhar, S.P.; Florante, R.; Cocca, S.M.; Editte, G. Saccharomyces cerevisiae Env7 is a novel serine/threonine kinase 16-related protein kinase and negatively regulates organelle fusion at the lysosomal vacuole. Mol. Cell. Biol. 2013, 33, 526–542. [Google Scholar] [CrossRef]
- Manandhar, S.P.; Calle, E.N.; Editte, G. Distinct palmitoylation events at the amino-terminal conserved cysteines of Env7 direct its stability, localization, and vacuolar fusion regulation in S. cerevisiae. J. Biol. Chem. 2014, 289, 11431–11442. [Google Scholar] [CrossRef]
- Nolen, B.; Taylor, S.; Ghosh, G. Regulation of Protein Kinases: Controlling Activity through Activation Segment Conformation. Mol. Cell 2004, 15, 661–675. [Google Scholar] [CrossRef]
- Natarajan, K.; Neuwald, A.F. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J. Mol. Biol. 2005, 351, 956–972. [Google Scholar]
- Kohn, A.D.; Summers, S.A.; Birnbaum, M.J.; Roth, R.A. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J. Biol. Chem. 1996, 271, 31372–31378. [Google Scholar] [CrossRef]
- Xiao, G.; Sun, S.C. Negative regulation of the nuclear factor κB-inducing kinase by a cis-acting domain. J. Biol. Chem. 2000, 275, 21081–21085. [Google Scholar] [CrossRef]
- Liu, J.; Sudom, A.; Min, X.; Cao, Z.; Gao, X.; Ayres, M.; Lee, F.; Cao, P.; Yehnston, S.; Plotnikova, O.; et al. Structure of the nuclear factor κB-inducing kinase (NIK) kinase domain reveals a constitutively active conformation. J. Biol. Chem. 2012, 287, 27326–27334. [Google Scholar] [CrossRef]
- Thu, Y.M.; Richmond, A. NF-κB inducing kinase: A key regulator in the immune system and in cancer. Cytokine Growth Factor Rev. 2010, 21, 213–226. [Google Scholar] [CrossRef]
- Zheng, W.; Cai, X.; Li, S.; Li, Z. Autophosphorylation mechanism of the Ser/Thr kinase Stk1 from Staphylococcus aureus. Front. Microbiol. 2018, 9, 758. [Google Scholar] [CrossRef]
- Romano, P.R.; Garcia-Barrio, M.T.; Zhang, X.; Wang, Q.; Taylor, D.R.; Zhang, F.; Herring, C.; Mathews, M.B.; Qin, J.; Hinnebusch, A.G. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol. Cell. Biol. 1998, 18, 2282–2297. [Google Scholar] [CrossRef]
- Saul, V.V.; Laureano, D.L.V.; Maja, M.; Marcus, K.; Thomas, B.; Karin, F.W.; Katja, B.; M Lienhard, S. HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J. Mol. Cell Biol. 2013, 5, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Stoffel, R.H.; Inglese, J.; Macrae, A.D.; Lefkowitz, R.J.; Premont, R.T. Palmitoylation increases the kinase activity of the G protein-coupled receptor kinase, GRK6. Biochemistry 1998, 37, 16053. [Google Scholar] [CrossRef]
- Mei, Y.; Wang, Y.; Hu, T.; Yang, X.; Lozano-Duran, R.; Sunter, G.; Zhou, X. Nucleocytoplasmic Shuttling of Geminivirus C4 Protein Mediated by Phosphorylation and Myristoylation Is Critical for Viral Pathogenicity. Mol. Plant 2018, 11, 1466–1481. [Google Scholar] [CrossRef]
- Kim, J.; Shishido, T.; Jiang, X.; Aderem, A.; Mclaughlin, S. Phosphorylation, high ionic strength, and calmodulin reverse the binding of MARCKS to phospholipid vesicles. J. Biol. Chem. 1994, 269, 28214–28219. [Google Scholar]
- Ohmori, S.; Sakai, N.; Shirai, Y.; Yamamoto, H.; Miyamoto, E.; Shimizu, N.; Saito, N. Importance of protein kinase C targeting for the phosphorylation of its substrate, myristoylated alanine-rich C-kinase substrate. J. Biol. Chem. 2000, 275, 26449–26457. [Google Scholar] [CrossRef]
- Hata, A.; Chen, Y.G. TGF-β Signaling from Receptors to Smads. Cold Spring Harb. Perspect. Biol. 2011, 8, a022061. [Google Scholar] [CrossRef]
- Macias, M.J.; Martin-Malpartida, P.; Massagué, J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem. Sci. 2015, 40, 296–308. [Google Scholar] [CrossRef]
- Heldin, C.H.; Moustakas, A. Signaling Receptors for TGF-β Family Members. Cold Spring Harb. Perspect. Biol. 2016, 8, a022053. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.E. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2017, 9, a022129. [Google Scholar] [CrossRef]
- Massagué, J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998, 67, 753–791. [Google Scholar] [CrossRef]
- Massagué, J.; Chen, Y.G. Controlling TGF-β signaling. Genes Dev. 2000, 14, 627–644. [Google Scholar]
- Rik, D.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar]
- Shi, Y.; Massague, J. Mechanisms of tgf-Beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Travis, M.A.; Sheppard, D. TGF-beta activation and function in immunity. Annu. Rev. Immunol. 2014, 32, 51–82. [Google Scholar] [CrossRef]
- Katz, L.H.; Ying, L.; Jiun-Sheng, C.; MuñOz, N.M.; Avijit, M.; Jian, C.; Lopa, M. Targeting TGF-β signaling in cancer. Expert Opin. Ther. Targets 2013, 17, 743. [Google Scholar] [CrossRef]
- Meng, X.M.; DJ, N.; Lan, H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325. [Google Scholar] [CrossRef]
- Ulrika, B.; Stefan, K. TGF-β signaling in the control of hematopoietic stem cells. Blood 2015, 125, 3542–3550. [Google Scholar]
- Li, J.; Wang, Y.; Meng, X.; Liang, H. Modulation of transcriptional activity in brain lower grade glioma by alternative splicing. PeerJ 2018, 6, e4686. [Google Scholar] [CrossRef] [Green Version]
- Park, J.T.; Uehara, T. How Bacteria Consume Their Own Exoskeletons (Turnover and Recycling of Cell Wall Peptidoglycan). Microbiol. Mol. Biol. Rev. 2008, 72, 211–227. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef] [Green Version]
- Moussian, B. The role of GlcNAc in formation and function of extracellular matrices. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 149, 215–226. [Google Scholar] [CrossRef]
- Dennis, J.W.; Nabi, I.R.; Demetriou, M. Metabolism, Cell Surface Organization, and Disease. Cell 2009, 139, 1229–1241. [Google Scholar] [CrossRef] [Green Version]
- Konopka, J.B. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo) 2012. [Google Scholar] [CrossRef]
- Wolosker, H.; Kline, D.; Bian, Y.; Blackshaw, S.; Cameron, A.M.; Fralich, T.J.; Schnaar, R.L.; Snyder, S.H. Molecularly cloned mammalian glucosamine-6-phosphate deaminase localizes to transporting epithelium and lacks oscillin activity. FASEB J. 1998, 12, 91–99. [Google Scholar] [CrossRef]
- Reutter, W.; Stäsche, R.; Stehling, P.; Baum, O. The Biology of Sialic Acids: Insights into their Structure, Metabolism and Function in Particular during Viral Infection. In Glycosciences: Status and Perspectives; Hans-Joachim, G., Sigrun, G., Eds.; Wiley: Hoboken, NJ, USA, 1997; pp. 245–259. [Google Scholar]
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 2015, 208, 869–880. [Google Scholar] [CrossRef]
- Corfield, A.P.; Berry, M. Glycan variation and evolution in the eukaryotes. Trends Biochem. Sci. 2015, 40, 351–359. [Google Scholar] [CrossRef]
- Shamoon, N.; Angelo, G.; Esteban, A.; Konopka, J.B. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J. Biol. Chem. 2011, 286, 28671. [Google Scholar]
- Thomas, N.; Joanne, H.; Mcconville, M.J. Evidence that intracellular stages of Leishmania major utilize amino sugars as a major carbon source. PLoS Pathog. 2010, 6, e1001245. [Google Scholar]
- Hyunsook, L.; Sun-Jung, C.; Il Soo, M. The non-canonical effect of N-acetyl-D-glucosamine kinase on the formation of neuronal dendrites. Mol. Cells 2014, 37, 248–256. [Google Scholar]
- Islam, M.A.; Sharif, S.R.; Lee, H.S.; Moon, I.S. N-Acetyl-D-Glucosamine Kinase Promotes the Axonal Growth of Developing Neurons. Mol. Cells 2015, 38, 876–885. [Google Scholar] [Green Version]
- Islam, M.A.; Sharif, S.R.; Lee, H.S.; Seog, D.H.; Moon, I.S. N-acetyl-D-glucosamine kinase interacts with dynein light-chain roadblock type 1 at Golgi outposts in neuronal dendritic branch points. Exp. Mol. Med. 2015, 47, e177. [Google Scholar] [CrossRef]
- Sharif, S.R.; Islam, A.; Moon, I.S. N-Acetyl-D-Glucosamine Kinase Interacts with Dynein-Lis1-NudE1 Complex and Regulates Cell Division. Mol. Cells 2016, 39, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Sharif, S.R.; Lee, H.; Islam, M.A.; Seog, D.H.; Moon, I.S. N-acetyl-D-glucosamine kinase is a component of nuclear speckles and paraspeckles. Mol. Cells 2015, 38, 402–408. [Google Scholar] [CrossRef]
- Wilson, S.H.; Bailey, A.M.; Nourse, C.R.; Mattei, M.G.; Byrne, J.A. Identification of MAL2, a novel member of the mal proteolipid family, though interactions with TPD52-like proteins in the yeast two-hybrid system. Genomics 2001, 76, 81–88. [Google Scholar] [CrossRef]
- De Marco, M.C.; Puertollano, R.; Martínezmenárguez, J.A.; Alonso, M.A. Dynamics of MAL2 during glycosylphosphatidylinositol-anchored protein transcytotic transport to the apical surface of hepatoma HepG2 cells. Traffic 2010, 7, 61–73. [Google Scholar] [CrossRef]
- In, J.G.; Tuma, P.L. MAL2 selectively regulates polymeric IgA receptor delivery from the Golgi to the plasma membrane in WIF-B cells. Traffic 2010, 11, 1056–1066. [Google Scholar] [CrossRef]
- Marco, M.C.; De Fernando, M.B.; Leonor, K.; Albar, J.P.; Isabel, C.; Vaerman, J.P.; Mónica, M.; Byrne, J.A.; Alonso, M.A. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J. Cell Biol. 2002, 159, 37–44. [Google Scholar] [CrossRef] [Green Version]
- In, J.G.; Striz, A.C.; Antonio, B.; Tuma, P.L. Serine/threonine kinase 16 and MAL2 regulate constitutive secretion of soluble cargo in hepatic cells. Biochem. J. 2014, 463, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Madrid, R.; Aranda, J.F.; Rodríguez-Fraticelli, A.E.; Ventimiglia, L.; Andrés-Delgado, L.; Shehata, M.; Fanayan, S.; Shahheydari, H.; Gómez, S.; Jiménez, A. The Formin INF2 Regulates Basolateral-to-Apical Transcytosis and Lumen Formation in Association with Cdc42 and MAL2. Dev. Cell 2010, 18, 814–827. [Google Scholar] [CrossRef] [Green Version]
- Rao, K.H.; Ghosh, S.; Datta, A. Env7p Associates with the Golgin Protein Imh1 at thetrans-Golgi Network inCandida albicans. MSphere 2016, 1, e00080-16. [Google Scholar] [CrossRef]
- Alfonso, L.C.; Striz, A.C.; Tuma, P.L. A Serine/Threonine Kinase 16-Based Phospho-Proteomics Screen Identifies WD Repeat Protein-1 as a Regulator of Constitutive Secretion. Sci. Rep. 2018, 8, 13049. [Google Scholar]
- Amberg, D.C.; Basart, E.; Botstein, D. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 1995, 2, 28–35. [Google Scholar] [CrossRef]
- Rodal, A.A.; Tetreault, J.W.; Lappalainen, P.; Drubin, D.G.; Amberg, D.C. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 1999, 145, 1251. [Google Scholar] [CrossRef]
- Shoichiro, O. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: New blades for twisted filaments. Biochemistry 2003, 42, 13363–13370. [Google Scholar]
- Gissen, P.; Arias, I.M. Structural and functional hepatocyte polarity and liver disease. J. Hepatol. 2015, 63, 1023–1037. [Google Scholar] [CrossRef] [Green Version]
- Zeigerer, A.; Wuttke, A.; Marsico, G.; Seifert, S.; Kalaidzidis, Y.; Zerial, M. Functional properties of hepatocytes in vitro are correlated with cell polarity maintenance. Exp. Cell Res. 2017, 350, 242–252. [Google Scholar] [CrossRef]
- Li, B.; Trueb, B. DRG represents a family of two closely related GTP-binding proteins. BBA-Gene Struct. Expr. 2000, 1491, 196–204. [Google Scholar] [CrossRef]
- Sazuka, T.; Kinoshita, M.; Tomooka, Y.; Ikawa, Y.; Noda, M.; Kumar, S. Expression of DRG during murine embryonic development. Biochem. Biophys. Res. Commun. 1992, 189, 371–377. [Google Scholar] [CrossRef]
- Sazuka, T.; Tomooka, Y.; Ikawa, Y.; Noda, M.; Kumar, S. DRG: A novel developmentally regulated GTP-binding protein. Biochem. Biophys. Res. Commun. 1992, 189, 363–370. [Google Scholar] [CrossRef]
- Mahajan, M.A.; Park, S.T.; Sun, X.H. Association of a novel GTP binding protein, DRG, with TAL oncogenic proteins. Oncogene 1996, 12, 2343–2350. [Google Scholar]
- Zhao, X.F.; Aplan, P.D. SCL binds the human homologue of DRG in vivo 1. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 1998, 1448, 109–114. [Google Scholar]
- Ishikawa, K.; Azuma, S.; Ikawa, S.; Morishita, Y.; Gohda, J.; Akiyama, T.; Semba, K.; Inoue, J.I. Cloning and characterization of Xenopus laevis drg2, a member of the developmentally regulated GTP-binding protein subfamily. Gene 2003, 322, 105–112. [Google Scholar] [CrossRef]
- Schellhaus, A.K.; Morenoandrés, D.; Chugh, M.; Yokoyama, H.; Moschopoulou, A.; De, S.; Bono, F.; Hipp, K.; Schäffer, E.; Antonin, W. Developmentally Regulated GTP binding protein 1 (DRG1) controls microtubule dynamics. Sci. Rep. 2017, 7, 9996. [Google Scholar] [CrossRef]
- Dang, T.; Jang, S.H.; Back, S.H.; Park, J.W.; Han, I.S. DRG2 Deficiency Causes Impaired Microtubule Dynamics in HeLa Cells. Mol. Cells 2018, 41, 1045–1051. [Google Scholar]
- Huang, S.; Wang, Y. Golgi structure formation, function, and post-translational modifications in mammalian cells. F1000Research 2017, 6, 2050. [Google Scholar] [CrossRef] [Green Version]
- Egea, G.; Lázaro-Diéguez, F.; Vilella, M. Actin dynamics at the Golgi complex in mammalian cells. Curr. Opin. Cell Biol. 2006, 18, 168–178. [Google Scholar] [CrossRef]
- Egea, G.; Serra-Peinado, C.; Salcedo-Sicilia, L.; Gutiérrez-Martínez, E. Actin acting at the Golgi. Histochem. Cell Biol. 2013, 140, 347–360. [Google Scholar] [CrossRef] [Green Version]
- Vinke, F.P.; Grieve, A.G.; Rabouille, C. The multiple facets of the Golgi reassembly stacking proteins. Biochem. J. 2011, 433, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Barr, F.A.; Puype, M.; Vandekerckhove, J.; Warren, G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 1997, 91, 253–262. [Google Scholar] [CrossRef]
- Tang, D.; Zhang, X.; Huang, S.; Yuan, H.; Li, J.; Wang, Y. Mena-GRASP65 interaction couples actin polymerization to Golgi ribbon linking. Mol. Biol. Cell 2016, 27, 137–152. [Google Scholar] [CrossRef]
- Romina Ines, C.; Raffaella, B.; Maria Luisa, B.; Daniela, S.; Inmaculada, A.; Nobuhiro, N.; Daniela, C.; Antonino, C. JNK2 controls fragmentation of the Golgi complex and the G2/M transition through phosphorylation of GRASP65. J. Cell Sci. 2015, 128, 2249–2260. [Google Scholar]
- Feinstein, T.N.; Linstedt, A.D. GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell 2008, 19, 2696–2707. [Google Scholar] [CrossRef]
- Valente, C.; Colanzi, A. Mechanisms and Regulation of the Mitotic Inheritance of the Golgi Complex. Front. Cell Dev. Biol. 2015, 3, 79. [Google Scholar] [CrossRef]
- Champion, L.; Linder, M.I.; Kutay, U. Cellular Reorganization during Mitotic Entry. Trends Cell Biol. 2016, 27, 26–41. [Google Scholar] [CrossRef]
- Lowe, M. Structural organization of the Golgi apparatus. Curr. Opin. Cell Biol. 2011, 23, 85–93. [Google Scholar] [CrossRef]
- Lin, C.M.; Chen, H.J.; Leung, C.L.; Parry, D.A.; Liem, R.K. Microtubule actin crosslinking factor 1b: A novel plakin that localizes to the Golgi complex. J. Cell Sci. 2005, 118, 3727–3738. [Google Scholar] [CrossRef]
- Wei, J.H.; Seemann, J. Golgi ribbon disassembly during mitosis, differentiation and disease progression. Curr. Opin. Cell Biol. 2017, 47, 43–51. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Ji, X.; Liu, J.; Zhang, X. Serine/Threonine Protein Kinase STK16. Int. J. Mol. Sci. 2019, 20, 1760. https://doi.org/10.3390/ijms20071760
Wang J, Ji X, Liu J, Zhang X. Serine/Threonine Protein Kinase STK16. International Journal of Molecular Sciences. 2019; 20(7):1760. https://doi.org/10.3390/ijms20071760
Chicago/Turabian StyleWang, Junjun, Xinmiao Ji, Juanjuan Liu, and Xin Zhang. 2019. "Serine/Threonine Protein Kinase STK16" International Journal of Molecular Sciences 20, no. 7: 1760. https://doi.org/10.3390/ijms20071760