Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Key Structural Conformations in MD Simulations
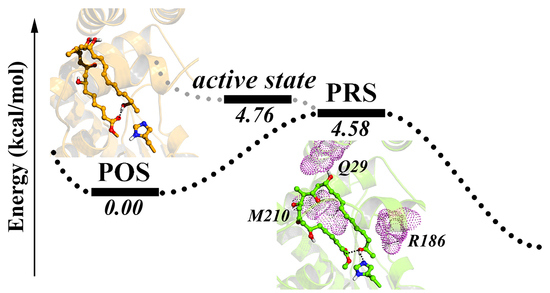
2.2. Conformational Transition between POS and PRS
2.3. Hydrophilic and Hydrophobic Interactions in Pima-TE System
2.4. Key Residues Analyzed Via Mutant Simulations
2.4.1. Mutation 1-Q29A
2.4.2. Mutation 2-M210G
2.4.3. Mutation 3-R186F & R186Y
2.4.4. Mutation 4-S138C
2.5. Study on TE’s Effect on the Release of Pimaricin Product
3. Discussion
4. Materials and Methods
4.1. System Preparation
4.2. Molecular Dynamics Simulation
4.3. Quantum Mechanics/Molecular Mechanics) Calculation
4.4. Simulation of Site Mutation Proteins
4.5. Free Energy Calculation and Conformational Stability Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PKS | Polyketide Synthase |
| TE | Thioesterase |
| MD | Molecular Dynamics |
| PRS | Pre-reaction State |
| POS | Pre-organization State |
References
- Gil, J.A.; Martin, J.F. Biotechnology of Antibiotics, 2nd ed.; W. Strohl, M., Ed.; Dekker: New York, NY, USA, 1997. [Google Scholar]
- Aparicio, J.F.; Mendes, M.V.; Anton, N.; Recio, E.; Martin, J.F. Polyene macrolide antibiotic biosynthesis. Curr. Med. Chem. 2004, 11, 1645–1656. [Google Scholar] [CrossRef]
- Szlinder-Richert, J.; Mazerski, J.; Cybulska, B.; Grzybowska, J.; Borowski, E. MFAME, N-methyl-N-d-fructosyl amphotericin B methyl ester, a new amphotericin B derivative of low toxicity: Relationship between self-association and effects on red blood cells. Biochim. Biophys. Acta Gen. Sub. 2001, 1528, 15–24. [Google Scholar] [CrossRef]
- Ogasawara, Y.; Katayama, K.; Minami, A.; Otsuka, M.; Eguchi, T.; Kakinuma, K. Cloning, Sequencing, and Functional Analysis of the Biosynthetic Gene Cluster of Macrolactam Antibiotic Vicenistatin in Streptomyces halstedii. Chem. Biol. 2006, 11, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Cereghetti, D.M.; Carreira, E.M. Amphotericin B: 50 Years of Chemistry and Biochemistry. Synthesis 2006, 37, 914–942. [Google Scholar] [CrossRef]
- Carmody, M.; Murphy, B.; Byrne, B.; Power, P.; Rai, D.; Rawlings, B.; Caffrey, P. Biosynthesis of Amphotericin Derivatives Lacking Exocyclic Carboxyl Groups. J. Biol. Chem. 2005, 280, 34420–34426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gantt, R.W.; Peltierpain, P.; Thorson, J.S. Enzymatic methods for glyco (diversification/randomization) of drugs and small molecules. Nat. Prod. Rep. 2011, 28, 1811–1853. [Google Scholar] [CrossRef]
- Zotchev, S.B. Polyene Macrolide Antibiotics and their Applications in Human Therapy. Curr. Med. Chem. 2003, 10, 211–223. [Google Scholar] [CrossRef]
- Baginski, M.; Czub, J.; Sternal, K. Interaction of amphotericin B and its selected derivatives with membranes: Molecular modeling studies. Chem. Rec. 2010, 6, 320–332. [Google Scholar] [CrossRef]
- Atta, H.M.; Selim, S.M.; Zayed, M.S. Natamycin antibiotic produced by Streptomyces sp.: Fermentation, purification and biological activities. J. Ame. Sci. 2012, 8, 469–475. [Google Scholar]
- Stark, J. Natamycin: An effective fungicide for food and beverages. Nat. Antimicrobials Minim. Process. Foods 2003, 82–97. [Google Scholar] [CrossRef]
- Austin, A.; Lietman, T.; Rose-nussbaumer, J. Update on the management of infectious keratitis. Ophthalmology 2017, 124, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
- Priya, A.B.; Kalyan, M. In vitro leishmanicidal effects of the anti-fungal drug natamycin are mediated through disruption of calcium homeostasis and mitochondrial dysfunction. Apoptosis 2018, 23, 420–435. [Google Scholar] [CrossRef]
- Te Welscher, Y.M.; Jones, L.; Van Leeuwen, M.R.; Dijksterhuis, J.; de Kruijff, B.; Eitzen, G.; Breukink, E. Natamycin Inhibits Vacuole Fusion at the Priming Phase via a Specific Interaction with Ergosterol. Antimicrob. Agents Chemother. 2010, 54, 2618–2625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanner, W. Membrane transport inhibition as mode of action of polyene antimycotics: Recent data supported by old ones. Food Technol. Biotechnol. 2014, 52, 8–12. [Google Scholar] [CrossRef]
- Van Leeuwen, M.R.; Golovina, E.A.; Dijksterhuis, J. The polyene antimycotics nystatin and filipin disrupt the plasma membrane, whereas natamycin inhibits endocytosis in germinating conidia of Penicillium discolor. J. Appl. Microbiol. 2009, 106, 1908–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mccall, L.I.; Aroussi, A.E.; Choi, J.Y.; Vieira, D.F.; Muylder, G.D.; Johnston, J.B.; Chen, S.; Kellar, D.; Siqueira-Neto, J.L.; Roush, W.R.; et al. Targeting Ergosterol Biosynthesis in Leishmania donovani: Essentiality of Sterol 14 alpha-demethylase. PLoS Negl. Trop. Dis. 2015, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Whicher, J.R.; Hansen, D.A.; Hansen, W.A.; Chelmer, J.A.; Congdon, G.R.; Alison, R.H.N.; Kristina, H.; Sherman, D.H.; Smith, J.L.; Skiniotis, G. Structure of a modular polyketide synthase. Nature 2014, 510, 512–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skiba, M.A.; Sikkema, A.P.; Fiers, W.D.; Gerwick, W.H.; Sherman, D.H.; Aldrich, C.C.; Smith, J.L. Domain Organization and Active Site Architecture of a Polyketide Synthase C-methyltransferase. ACS Chem. Biol. 2016, 11, 3319–3327. [Google Scholar] [CrossRef]
- Curran, S.C.; Hagen, A.; Poust, S.; Chan, L.J.G.; Garabedian, B.M.; Rond, T.; Baluyot, M.J.; Vu, J.T.; Lau, A.K.; Yuzawa, S.; et al. Probing the flexibility of an iterative modular polyketide synthase with non-native substrates in vitro. ACS Chem. Biol. 2018, 13, 2261–2268. [Google Scholar] [CrossRef]
- Rittner, A.; Paithankar, K.S.; Vu, K.H.; Grininger, M. Characterization of the polyspecific transferase of murine type I fatty acid synthase (FAS) and implications for polyketide synthase (PKS) engineering. ACS Chem. Biol. 2018, 13, 723–732. [Google Scholar] [CrossRef]
- Ferscht, A. Enzyme Structure and Mechanism, 2nd ed.; W. H. Freeman and Company: New York, NY, USA, 1985. [Google Scholar] [CrossRef]
- Kormana, T.P.; Crawfordb, J.M.; Labonteb, J.W.; Newmanb, A.G.; Wongc, J.; Townsendb, C.A.; Tsaia, S.C. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 6246–6251. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.P.; Shi, T.; Wang, X.L.; Wang, J.T.; Chen, Q.H.; Bai, L.Q.; Zhao, Y.L. Theoretical studies on the Mechanism of Thioesterase-catalyzed Macrocyclization in Erythromycin Biosynthesis. ACS Catal. 2016, 6, 4369–4378. [Google Scholar] [CrossRef]
- Trauger, J.W.; Kohli, R.M.; Walsh, C.T. Cyclization of Backbone-Substituted Peptides Catalyzed by the Thioesterase Domain from the Tyrocidine Nonribosomal Peptide Synthetase. Biochemistry 2001, 40, 7092–7098. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, S.; Murthy, M.R.N. Analysis of temperature factor distribution in high-resolution protein structures. Protein Sci. 1997, 6, 2561–2567. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.Y.; Shen, H.B. Robust Prediction of B-Factor Profile from Sequence Using Two-Stage SVR Based on Random Forest Feature Selection. Protein Peptide Lett. 2009, 16, 1447–1454. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Koch, A.A.; Hansen, D.A.; Shende, V.V.; Furan, L.R.; Houk, K.N.; Gonzalo Jiménez-Osés, G.; Sherman., D.H. A Single Active Site Mutation in the Pikromycin Thioesterase Generates a More Effective Macrocyclization Catalyst. J. Am. Chem. Soc. 2017, 139, 13456–13465. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F.W. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Onur, S.; Pemra, O. gRINN: A tool for calculation of residue interaction energies and protein energy network analysis of molecular dynamics simulations. Nucleic Acids Res. 2018, 46, W554–W562. [Google Scholar] [CrossRef]
- Li, J.; Sun, R.; Wu, Y.H.; Song, M.Z.; Li, J.; Yang, Q.Y.; Chen, X.Y.; Bao, J.K.; Zhao, Q. L1198F Mutation Resensitizes Crizotinib to ALK by Altering the Conformation of Inhibitor and ATP Binding Sites. Int. J. Mol. Sci. 2017, 18, 482. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Gokey, T.; Ting, D.; He, Z.H.; Guliaev, A.B. Dimerization misalignment in human glutamate-oxaloacetate transaminase variants is the primary factor for PLP release. PLOS ONE 2018, 13, e0203889. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.P.; Liu, G.J.; Zhou, H.Y.; Fang, X.; Fang, Y.; Wu, J.H. Computer prediction of paratope on antithrombotic antibody 10B12 and epitope on platelet glycoprotein VI via molecular dynamics simulation. BioMed. Eng. OnLine 2016, 15, 647–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, T.; Liu, L.X.; Tao, W.T.; Luo, S.G.; Fan, S.B.; Wang, X.L.; Bai, L.Q.; Zhao, Y.L. Theoretical studies on the Catalytic Mechanism and Substrate Diversity for Macrocyclization of Pikromycin Thioesterase. ACS Catal. 2018, 8, 4323–4332. [Google Scholar] [CrossRef]
- Giraldes, J.W.; Akey, D.L.; Kittendorf, J.D.; Sherman, D.H.; Smith, J.L.; Fecik, R.A. Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels. Nat. Chem. Biol. 2006, 2, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Accelrys Discovery Studio Visualizer 3.5; Accelerys Software Inc.: San Diego, CA, USA, 2005.
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Amber 2014; Univeristy of California: San Francisco, CA, USA, 2014.
- Jakalian, A.; Bush, B.L.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic Charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000, 21, 132–146. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Gaussian 09; Revision A.02; Gaussian Inc.: Wallingford, CT, USA, 2009.
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.M.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. J. Chem. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Vreven, T.; Byun, K.S.; Komáromi, I.; Dapprich, S.; Montgomery, J.A., Jr.; Morokuma, K.; Frisch, M.J. Combining quantum mechanics methods with molecular mechanics methods in ONIOM. J. Chem. Theory Comput. 2006, 2, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Vreven, T.; Frisch, M.; Kudin, K.; Schlegel, H.; Morokuma, K. Geometry optimization with QM/MM methods II: Explicit quadratic coupling. Mol. Phys. 2006, 104, 701–714. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Rassolov, V.A.; Ratner, M.A.; Pople, J.A.; Redfern, P.C.; Curtiss, L.A. 6-31G* Basis Set for Third-Row Atoms. J. Comput. Chem. 2001, 22, 976–984. [Google Scholar] [CrossRef]
- Swanson, J.M.J.; Henchman, R.H.; McCammon, J.A. Revisiting free energy calculations: A theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophys. J. 2004, 86, 67–74. [Google Scholar] [CrossRef]











| Substrate Type | Name | System | No. of Runs Per Complex | Length Per Run (ns) |
|---|---|---|---|---|
| Polyketide Chain | wild type | pima-TEWT + polyketide chain | 5 | 50 |
| M210G | pima-TEM210G + polyketide chain | 3 | 30 | |
| Q29A | pima-TEQ29A + polyketide chain | 3 | 30 | |
| R186F | pima-TER186F + polyketide chain | 3 | 30 | |
| R186Y | pima-TER186Y + polyketide chain | 3 | 30 | |
| S138C | pima-TES138C + polyketide chain | 3 | 50 | |
| Product | ring | pima-TEWT + product | 3 | 50 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Wang, R.; Li, C.; Bai, L.; Zhao, Y.-L.; Shi, T. Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase. Int. J. Mol. Sci. 2019, 20, 877. https://doi.org/10.3390/ijms20040877
Fan S, Wang R, Li C, Bai L, Zhao Y-L, Shi T. Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase. International Journal of Molecular Sciences. 2019; 20(4):877. https://doi.org/10.3390/ijms20040877
Chicago/Turabian StyleFan, Shuobing, Rufan Wang, Chen Li, Linquan Bai, Yi-Lei Zhao, and Ting Shi. 2019. "Insight into Structural Characteristics of Protein-Substrate Interaction in Pimaricin Thioesterase" International Journal of Molecular Sciences 20, no. 4: 877. https://doi.org/10.3390/ijms20040877