Clustered DNA Damages induced by 0.5 to 30 eV Electrons
Abstract
:1. Introduction
1.1. Clustered DNA Damages and Relationship to Cell Survival
1.2. Clustered Damages Induced by Ionizing Radiation
1.3. Role of LEEs
2. Experimental Techniques
2.1. Target Preparation and LEE Irradiation
2.2. Analysis of DNA Conformation Variations and Cell Survival
2.3. Enzyme Treatment for Measuring Base Damages
3. Clustered DNA Damages Induced by LEEs
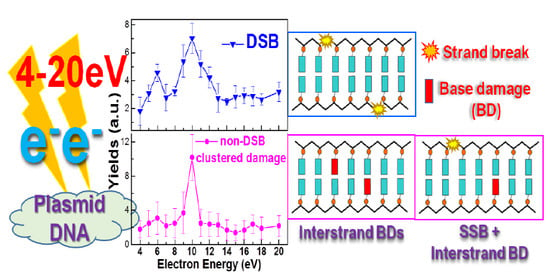
3.1. DSBs
3.2. Clustered Lesions Involving Base Damage
4. Cross-Sections of Clustered DNA Damages
5. Enhancement of Clustered DNA Damages by Chemotherapeutic Agents and Radiosensitizers
6. Mechanism of clustered DNA damage induced by a single LEE
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| A | adenine |
| AP | Abasic site (apurinic or apyrimidinic) |
| BD | base damage |
| BER | base excision repair |
| bp | base pair |
| C | cytosine |
| CA | chemotherapeutic agent |
| CRT | chemoradiation therapy |
| CS | cross section |
| DEA | dissociative electron attachment |
| DNA | deoxyribonucleic acid |
| DSB | double strand break |
| EF | enhancement factor |
| f | penetration factor |
| Fpg | formamidopyrimidine N-glycosylase |
| G | guanine |
| IR | ionizing radiation |
| LEE | low-energy electron |
| LET | linear energy transfer |
| LMDS | locally multiple damage sites |
| MC | Monte Carlo |
| Nth | base excision repair endonuclease III |
| SSB | single strand break |
| T | thymine |
| TA | transient anions |
| U | uracil |
References
- Eccles, L.; O’Neill, P.; Lomax, M. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res. 2011, 711, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Ravanat, J.-L.; TavernaPorro, M.; Menoni, H.; Angelov, D. Oxidatively generated complex DNA damage: Tandem and clustered lesions. Cancer Lett. 2012, 327, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Gulston, M.; De Lara, C.; Jenner, T.; Davis, E.; O’Neill, P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004, 32, 1602–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kejnovská, I.; Bednárová, K.; Renciuk, D.; Dvoráková, Z.; Školáková, P.; Trantírek, L.; Fiala, R.; Vorlícková, M.; Sagi, J. Clustered abasic lesions profoundly change the structure and stability of human telomeric G-quadruplexes. Nucleic Acids Res. 2017, 45, 4294–4305. [Google Scholar] [CrossRef] [PubMed]
- Shikazono, N.; Akamatsu, K. Mutagenic potential of 8-oxo-7,8-dihydroguanine (8-oxoG) is influenced by nearby clustered lesions. Mutat. Res. Mol. Mech. Mutagen. 2018, 810, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Bignon, E.; Gattuso, H.; Morell, C.; Dehez, F.; Georgakilas, A.G.; Monari, A.; Dumont, E. Correlation of bistranded clustered abasic DNA lesion processing with structural and dynamic DNA helix distortion. Nucleic Acids Res. 2016, 44, 8588–8599. [Google Scholar] [CrossRef] [Green Version]
- Sage, E.; Harrison, L. Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat. Res. 2011, 711, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [Green Version]
- Bukowska, B.; Karwowski, B.T. The clustered DNA lesions—Types, pathways of repair and relevance to human health. Curr. Med. Chem. 2018, 25, 2722–2735. [Google Scholar] [CrossRef]
- Ward, J. The complexity of DNA damage: Relevance to biological consequences. Int. J. Radiat. Biol. 1994, 66, 427–432. [Google Scholar] [CrossRef]
- Hill, M. Track to the future: Historical perspective on the importance of radiation track structure and DNA as a radiobiological target. Int. J. Radiat. Biol. 2018, 94, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Fuciarelli, A.F.; Zimbrick, J.D. (Eds.) Radiation Damage in DNA: Structure/Function Relationships at Early Times; Battelle: Columbus, OH, USA, 1995. [Google Scholar]
- Nikitaki, Z.; Nikolov, V.; Mavragani, I.V.; Plante, I.; Emfietzoglou, D.; Iliakis, G.; Georgakilas, A.G. Non-DSB clustered DNA lesions. Does theory colocalize with the experiment? Radiat. Phys. Chem. 2016, 128, 26–35. [Google Scholar] [CrossRef]
- Ward, J.F. Radiation mutagenesis: The initial DNA lesions responsible. Radiat. Res. 1995, 142, 362. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, B.M.; Bennett, P.V.; Sidorkina, O.; Laval, J. Clustered damages and total lesions induced in DNA by ionizing radiation: Oxidized bases and strand breaks. Biochemistry 2000, 39, 8026–8031. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, B.M.; Bennett, P.V.; Sutherland, J.C.; Laval, J. Clustered DNA damages induced by X rays in human cells. Radiat. Res. 2002, 157, 611–616. [Google Scholar] [CrossRef]
- Islam, M.T. Radiation interactions with biological system. Int. J. Radiat. Biol. 2017, 93, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Timm, S.; Lorat, Y.; Jakob, B.; Taucher-Scholz, G.; Rübe, C.E. Clustered DNA damage concentrated in particle trajectories causes persistent large-scale rearrangements in chromatin architecture. Radiother. Oncol. 2018, 129, 600–610. [Google Scholar] [CrossRef]
- Sutherland, B.; Bennett, P.; Sidorkina, O.; Laval, J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. USA 2000, 97, 103–108. [Google Scholar] [CrossRef] [Green Version]
- Gulston, M.; Fulford, J.; Jenner, T.; De Lara, C.; O’Neill, P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002, 30, 3464–3472. [Google Scholar] [CrossRef] [Green Version]
- Monteleone, J.; Sutherland, B. Clustered DNA damages induced by high and low LET radiation, including heavy ions. Phys. Med. 2001, 17 (Suppl. 1), 202–204. [Google Scholar]
- Tsao, D.; Kalogerinis, P.; Tabrizi, I.; Dingfelder, M.; Stewart, R.; Georgakilas, A. Induction and processing of oxidative clustered DNA lesions in 56Fe-ionirradiated human monocytes. Radiat. Res. 2007, 168, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, Y.; Furusawa, Y.; Ide, H.; Yasui, A.; Terato, H. Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. J. Radiat. Res. 2015, 56, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, I.; Shikazono, N.; Suzuki, M.; Fujii, K.; Yokoya, A. Efficiency of radiation-induced base lesion excision and the order of enzymatic treatment. Int. J. Radiat. Biol. 2017, 93, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Jenner, T.J.; Fulford, J.; O’Neill, P. Contribution of base lesions to radiation-induced clustered DNA damage: Implication for models of radiation response. Radiat. Res. 2001, 156, 590–593. [Google Scholar] [CrossRef]
- Hada, M.; Georgakilas, A.G. Formation of clustered DNA damage after high-LET irradiation: A review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Shikazono, N.; Noguchi, M.; Fujii, K.; Urushibara, A.; Yokoya, A. The yield, processing, and biological consequences of clustered DNA damage induced by ionizing radiation. J. Radiat. Res. 2009, 50, 27–36. [Google Scholar] [CrossRef]
- Georgakilas, A.; O’Neill, P.; Stewart, R. Induction and repair of clustered DNA lesions: What do we know so far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef]
- Sage, E.; Shikazono, N. Radiation-induced clustered DNA lesions: Repair and mutagenesis. Free Radic. Biol. Med. 2017, 107, 125–135. [Google Scholar] [CrossRef]
- Lampe, N.; Karamitros, M.; Breton, V.; Brown, J.M.; Kyriakou, I.; Sakata, D.; Sarramia, D.; Incerti, S. Mechanistic DNA damage simulations in Geant4-DNA part 1: A parameter study in a simplified geometry. Phys. Med. 2018, 48, 135–145. [Google Scholar] [CrossRef]
- Buch, T.; Scifoni, E.; Durante, M.; Scholz, M.; Kramer, M.; Friedrich, T. Modeling radiation effects of ultrasoft X rays on the basis of amorphous track structure. Radiat. Res. 2018, 189, 32–43. [Google Scholar] [CrossRef]
- Nikjoo, H.; O’Neill, P.; Goodhead, D.T.; Terrissol, M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997, 71, 467–483. [Google Scholar] [CrossRef] [PubMed]
- Nikjoo, H.; Uehara, S.; Wilson, W.E.; Hoshi, M.; Goodhead, D.T. Track structure in radiation biology: Theory and applications. Int. J. Radiat. Biol. 1998, 73, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Nikjoo, H.; O’Neill, P.; Terrissol, M.; Goodhead, D.T. Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat. Environ. Biophys. 1999, 38, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Nikjoo, H.; O’Neill, P.; Wilson, W.E.; Goodhead, D.T. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 2001, 156, 577–583. [Google Scholar] [CrossRef]
- Taleei, R.; Girard, P.M.; Sankaranarayanan, K.; Nikjoo, H. The non-homologous end-joining (NHEJ) mathematical model for the repair of double-strand breaks: II. Application to damage induced by ultrasoft X rays and low-energy electrons. Radiat. Res. 2013, 179, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Nikjoo, H.; Emfietzoglou, D.; Liamsuwan, T.; Taleei, R.; Liljequist, D.; Uehara, S. Radiation track, DNA damage and response—A review. Rep. Prog. Phys. 2016, 79, 116601. [Google Scholar] [CrossRef] [PubMed]
- Streitmatter, S.W.; Stewart, R.D.; Jenkins, P.A.; Jevremovic, T. DNA double strand break (DSB) induction and cell survival in iodine-enhanced computed tomography (CT). Phys. Med. Biol. 2017, 62, 6164–6184. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tan, Z.; Zhang, L.; Champion, C. Investigation on the correlation between energy deposition and clustered DNA damage induced by low-energy electrons. Radiat. Environ. Biophys. 2018, 57, 179–187. [Google Scholar] [CrossRef]
- Kai, T.; Yokoya, A.; Ukai, M.; Fujii, K.; Toigawa, T.; Watanabe, R. A significant role of non-thermal equilibrated electrons in the formation of deleterious complex DNA damage. Phys. Chem. Chem. Phys. 2018, 20, 2838–2844. [Google Scholar] [CrossRef]
- Brenner, D.; Ward, J. Constraints on energy deposition and target size of multiply damaged sites associated with DNA double-strand breaks. Int. J. Radiat. Biol. 1992, 61, 737–748. [Google Scholar] [CrossRef]
- Botchway, S.W.; Stevens, D.L.; Hill, M.A.; Jenner, T.J.; O’Neill, P. Induction and rejoining of DNA double-strand breaks in Chinese hamster V79-4 cells irradiated with characteristic aluminum K and copper L ultrasoft X rays. Radiat. Res. 1997, 148, 317. [Google Scholar] [CrossRef]
- Olive, P.L. The role of DNA single- and double-strand breaks in cell killing by ionizing radiation. Radiat. Res. 1998, 150, S42. [Google Scholar] [CrossRef]
- Rübe, C.E.; Dong, X.; Kühne, M.; Fricke, A.; Kaestner, L.; Lipp, P.; Rübe, C. DNA double-strand break rejoining in complex normal tissues. Int. J. Radiat. Oncol. 2008, 72, 1180–1187. [Google Scholar] [CrossRef] [PubMed]
- Lobrich, M.; Shibata, A.; Beucher, A.; Fisher, A.; Ensminger, M.; Goodarzi, A.; Barton, O.; Jeggo, P. Gamma H2AX foci analysis for monitoring DNA double strand break repair: Strengths, limitations and optimization. Cell Cycle 2010, 9, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.H. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutat. Res. 2012, 751, 158–246. [Google Scholar] [CrossRef] [PubMed]
- Povirk, L.F. Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol. Biol. 2012, 2012, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Schipler, A.; Iliakis, G. DNA double-strand–break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef] [PubMed]
- Leatherbarrow, E.; Harper, J.; Cucinotta, F.; O’Neill, P. Induction and quantification of gamma-H2AX foci following low and high LET-irradiation. Int. J. Radiat. Biol. 2006, 82, 111–118. [Google Scholar] [CrossRef]
- Von Sonntag, C. The Chemical Basis of Radiation Biology; Taylor & Francis: London, UK, 1987. [Google Scholar]
- Cadet, J.; Wagner, R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef]
- David-Cordonnier, M.-H. Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein. Nucleic Acids Res. 2001, 29, 1107–1113. [Google Scholar] [CrossRef] [Green Version]
- Song, J.M.; Milligan, J.R.; Sutherland, B.M. Bistranded oxidized purine damage clusters: Induced in DNA by long-wavelength ultraviolet (290–400 nm) radiation? Biochemistry 2002, 41, 8683–8688. [Google Scholar] [CrossRef] [PubMed]
- Milligan, J.R.; Aguilera, J.A.; Nguyen, T.T.; Paglinawan, R.A.; Ward, J.F. DNA strand-break yields after post-irradiation incubation with base excision repair endonucleases implicate hydroxyl radical pairs in double-strand break formation. Int. J. Radiat. Biol. 2000, 76, 1475–1483. [Google Scholar] [PubMed]
- Milligan, J.; Aguilera, J.A.; Paglinawan, R.A.; Ward, J.F.; Limoli, C.L.; Milligan, J.R. DNA strand break yields after post-high LET irradiation incubation with endonuclease-III and evidence for hydroxyl radical clustering. Int. J. Radiat. Biol. 2001, 77, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Yokoya, A.; Cunniffe, S.M.T.; O’Neill, P. Effect of hydration on the induction of strand breaks and base lesions in plasmid DNA films by γ-radiation. J. Am. Chem. Soc. 2002, 124, 8859–8866. [Google Scholar] [CrossRef] [PubMed]
- Yokoya, A.; Cunniffe, S.M.T.; Watanabe, R.; Kobayashi, K.; O’Neill, P. Induction of DNA strand breaks, base lesions and clustered damage sites in hydrated plasmid DNA films by ultrasoft X Rays around the phosphorus K edge. Radiat. Res. 2009, 172, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, B.M.; Bennett, P.V.; Cintron-Torres, N.; Hada, M.; Trunk, J.; Monteleone, D.; Sutherland, J.C.; Laval, J.; Stanislaus, M.; Gewirtz, A. Clustered DNA damages induced in human hematopoietic cells by low doses of ionizing radiation. J. Radiat. Res. 2002, 43, S149–S152. [Google Scholar] [CrossRef]
- David-Cordonnier, M.-H. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem. 2000, 275, 11865–11873. [Google Scholar] [CrossRef]
- Pimblott, S.M.; LaVerne, J.A. Production of low-energy electrons by ionizing radiation. Radiat. Phys. Chem. 2007, 76, 1244–1247. [Google Scholar] [CrossRef]
- Alizadeh, E.; Sanche, L. Precursors of Solvated Electrons in Radiation Biology. Chem. Rev. 2012, 112, 5578–5602. [Google Scholar] [CrossRef]
- Arumainayagam, C.R.; Lee, H.-L.; Nelson, R.B.; Haines, D.R.; Gunawardane, R.P. Low-energy electron-induced reactions in condensed matter. Surf. Sci. Rep. 2010, 65, 1–44. [Google Scholar] [CrossRef]
- Böhler, E.; Warneke, J.; Swiderek, P. Control of chemical reactions and synthesis by low-energy electrons. Chem. Soc. Rev. 2013, 42, 9219. [Google Scholar]
- Trinter, F.; Schöffler, M.S.; Kim, H.K.; Sturm, F.P.; Cole, K.; Neumann, N.; Vredenborg, A.; Williams, J.; Bocharova, I.; Guillemin, R.; et al. Resonant Auger decay driving intermolecular Coulombic decay in molecular dimers. Nature 2014, 505, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Sanche, L. Low-energy electron interaction with DNA: Bond dissociation and formation of transient anions, radicals, and radical anions. In Radical and Radical Ion Reactivity in Nucleic Acid Chemistry; Wiley: Hoboken, NJ, USA, 2009; pp. 239–293. [Google Scholar]
- Alizadeh, E.; Orlando, T.M.; Sanche, L. Biomolecular damage induced by ionizing radiation: The direct and indirect effects of low-energy electrons on DNA. Annu. Rev. Phys. Chem. 2015, 66, 379–398. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Gao, Y.; Liu, W.; Gao, T.; Zheng, Y.; Sanche, L. Clustered DNA damages induced by 2–20 eV electrons and transient anions: General mechanism and correlation to cell death. J. Phys. Chem. Lett. 2019, 10, 2985–2990. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, M.; Hill, R.P.; Jaffray, D.A. The exploitation of low-energy electrons in cancer treatment. Radiat. Res. 2017, 188, 123–143. [Google Scholar] [CrossRef] [PubMed]
- Schürmann, R.; Vogel, S.; Ebel, K.; Bald, I. Frontispiece: The physico-chemical basis of DNA radiosensitization: Implications for cancer radiation therapy. Chem. Eur. J. 2018, 24, 10271–10279. [Google Scholar] [CrossRef] [PubMed]
- Gorfinkiel, J.D.; Ptasinska, S. Electron scattering from molecules and molecular aggregates of biological relevance. J. Phys. B Mol. Opt. Phys. 2017, 50, 182001. [Google Scholar] [CrossRef]
- Baccarelli, I.; Bald, I.; Gianturco, F.A.; Illenberger, E.; Kopyra, J. Electron-induced damage of DNA and its components: Experiments and theoretical models. Phys. Rep. 2011, 508, 1–44. [Google Scholar] [CrossRef]
- Alizadeh, E.; Ptasińska, S.; Sanche, L. Transient anions in radiobiology and radiotherapy: From gaseous biomolecules to condensed organic and biomolecular solids. In Radiation Effects in Materials; Monteiro, W.A., Ed.; Intech Open: London, UK, 2016. [Google Scholar]
- Matthews, E.; Cercola, R.; Mensa-Bonsu, G.; Neumark, D.M.; Dessent, C.E.H. Photoexcitation of iodide ion-pyrimidine clusters above the electron detachment threshold: Intracluster electron transfer versus nucleobase-centred excitations. J. Chem. Phys. 2018, 148, 084304. [Google Scholar] [CrossRef]
- Kočišek, J.; Sedmidubská, B.; Indrajith, S.; Fárník, M.; Fedor, J. Electron attachment to microhydrated deoxycytidine monophosphate. J. Phys. Chem. B 2018, 122, 5212–5217. [Google Scholar] [CrossRef]
- Ma, J.; Denisov, S.A.; Marignier, J.-L.; Pernot, P.; Adhikary, A.; Seki, S.; Mostafavi, M. Ultrafast electron attachment and hole transfer following ionizing radiation of aqueous uridine monophosphate. J. Phys. Chem. Lett. 2018, 9, 5105–5109. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Kumar, A.; Muroya, Y.; Yamashita, S.; Sakurai, T.; Denisov, S.A.; Sevilla, M.D.; Adhikary, A.; Seki, S.; Mostafavi, M. Observation of dissociative quasi-free electron attachment to nucleoside via excited anion radical in solution. Nat. Commun. 2019, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Ptasińska, S.; Sanche, L. On the mechanism of anion desorption from DNA induced by low energy electrons. J. Chem. Phys. 2006, 125, 144713. [Google Scholar]
- McKee, A.D.; Schaible, M.J.; Rosenberg, R.A.; Kundu, S.; Orlando, T.M. Low energy secondary electron induced damage of condensed nucleotides. J. Chem. Phys. 2019, 150, 204709. [Google Scholar] [CrossRef] [PubMed]
- Schürmann, R.; Tsering, T.; Tanzer, K.; Denifl, S.; Kumar, S.V.K.; Bald, I. Resonant formation of strand breaks in sensitized oligonucleotides induced by low-energy electrons (0.5–9 eV). Angew. Chem. Int. Ed. 2017, 56, 10952–10955. [Google Scholar] [CrossRef] [PubMed]
- Kunin, A.; Neumark, D.M. Time-resolved radiation chemistry: Femtosecond photoelectron spectroscopy of electron attachment and photodissociation dynamics in iodide-nucleobase clusters. Phys. Chem. Chem. Phys. 2019, 21, 7239–7255. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.B.; Meyer, S.; Schröter, M.-A.; Seitz, H.; Kunte, H.-J.; Solomun, T.; Sturm, H. Direct electron irradiation of DNA in a fully aqueous environment. Damage determination in combination with Monte Carlo simulations. Phys. Chem. Chem. Phys. 2017, 19, 1798–1805. [Google Scholar] [CrossRef]
- Meesat, R.; Belmouaddine, H.; Allard, J.-F.; Tanguay-Renaud, C.; Lemay, R.; Brastaviceanu, T.; Tremblay, L.; Paquette, B.; Wagner, J.R.; Jay-Gerin, J.-P.; et al. Cancer radiotherapy based on femtosecond IR laser-beam filamentation yielding ultra-high dose rates and zero entrance dose. Proc. Natl. Acad. Sci. USA 2012, 109, E2508–E2513. [Google Scholar] [CrossRef]
- Belmouaddine, H.; Madugundu, G.S.; Wagner, J.R.; Couairon, A.; Houde, D.; Sanche, L. DNA base modifications mediated by femtosecond laser-induced cold low-density plasma in aqueous solutions. J. Phys. Chem. Lett. 2019, 10, 2753–2760. [Google Scholar] [CrossRef]
- Liang, X.-X.; Zhang, Z.; Vogel, A. Multi-rate-equation modeling of the energy spectrum of laser-induced conduction band electrons in water. Opt. Express 2019, 27, 4672–4693. [Google Scholar] [CrossRef]
- Yokoya, A.; Ito, T. Photon-induced auger effect in biological systems: A review. Int. J. Radiat. Biol. 2017, 93, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zheng, Y.; Sanche, L. DNA strand breaks and crosslinks induced by transient anions in the range 2–20 eV. J. Chem. Phys. 2014, 140, 155101. [Google Scholar] [CrossRef] [PubMed]
- Boulanouar, O.; Khatyr, A.; Herlem, G.; Palmino, F.; Sanche, L.; Fromm, M. Soft adsorption of densely packed layers of DNA-plasmid·1,3-diaminopropane complexes onto highly oriented pyrolitic graphite designed to erode in water. J. Phys. Chem. C 2011, 115, 21291–21298. [Google Scholar] [CrossRef]
- Nikitaki, Z.; Nikolov, V.; Mavragani, I.V.; Mladenov, E.; Mangelis, A.; Laskaratou, D.A.; Fragkoulis, G.I.; Hellweg, C.E.; Martin, O.A.; Emfietzoglou, D.; et al. Measurement of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET). Free Radic. Res. 2016, 50, S64–S78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahbani, S.K.; Sanche, L.; Cloutier, P.; Bass, A.D.; Hunting, D.J. Loss of cellular transformation efficiency induced by DNA irradiation with low-energy (10 eV) electrons. J. Phys. Chem. B 2014, 118, 13123–13131. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. DNA glycosylases search for and remove oxidized DNA bases. Environ. Mol. Mutagen. 2013, 54, 691–704. [Google Scholar] [CrossRef] [Green Version]
- Burrows, C.J.; Muller, J.G. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 2009, 109, 2929–2950. [Google Scholar] [CrossRef]
- Shikazono, N.; Akamatsu, K.; Takahashi, M.; Noguchi, M.; Urushibara, A.; O’Neill, P.; Yokoya, A. Significance of DNA polymerase I in in vivo processing of clustered DNA damage. Mutat. Res. Mol. Mech. Mutagen. 2013, 749, 9–15. [Google Scholar] [CrossRef]
- Lee, A.J.; Wallace, S.S. Visualizing the search for radiation-damaged DNA bases in real time. Radiat. Phys. Chem. 2016, 128, 126–133. [Google Scholar] [CrossRef] [Green Version]
- Folkard, M.; Prise, K.M.; Vojnovic, B.; Davies, S.; Roper, M.J.; Michael, B.D. Measurement of DNA damage by electrons with energies between 25 and 4000 eV. Int. J. Radiat. Biol. 1993, 64, 651–658. [Google Scholar] [CrossRef]
- Huels, M.A.; Boudaïffa, B.; Cloutier, P.; Hunting, D.; Sanche, L. Single, double, and multiple double strand breaks induced in DNA by 3–100 eV electrons. J. Am. Chem. Soc. 2003, 125, 4467–4477. [Google Scholar] [CrossRef] [PubMed]
- Orlando, T.M.; Oh, D.; Chen, Y.; Aleksandrov, A.B. Low-energy electron diffraction and induced damage in hydrated DNA. J. Chem. Phys. 2008, 128, 195102. [Google Scholar] [CrossRef]
- Ptasinska, S.; Sanche, L. Dissociative electron attachment to hydrated single DNA strands. Phys. Rev. E 2007, 75, 031915. [Google Scholar] [CrossRef]
- Cai, Z.; Cloutier, P.; Hunting, D.; Sanche, L. Comparison between X-ray photon and secondary electron damage to DNA in vacuum. J. Phys. Chem. B 2005, 109, 4796–4800. [Google Scholar] [CrossRef]
- Alizadeh, E.; Sanche, L. Role of humidity and oxygen level on damage to DNA induced by soft X-rays and low-energy electrons. J. Phys. Chem. C 2013, 117, 22445–22453. [Google Scholar] [CrossRef] [PubMed]
- Sahbani, S.K.; Cloutier, P.; Bass, A.D.; Hunting, D.J.; Sanche, L. Electron resonance decay into a biological function: Decrease in viability of E. coli transformed by plasmid DNA irradiated with 0.5–18 eV electrons. J. Phys. Chem. Lett. 2015, 6, 3911–3914. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Sanche, L. Effective and absolute cross sections for low-energy (1–30 eV) electron interactions with condensed biomolecules. Appl. Phys. Rev. 2018, 5, 021302. [Google Scholar] [CrossRef]
- Rezaee, M.; Cloutier, P.; Bass, A.D.; Michaud, M.; Hunting, D.J.; Sanche, L. Absolute cross section for low-energy-electron damage to condensed macromolecules: A case study of DNA. Phys. Rev. E 2012, 86, 031913. [Google Scholar] [CrossRef]
- Chen, W.; Chen, S.; Dong, Y.; Cloutier, P.; Zheng, Y.; Sanche, L. Absolute cross-sections for DNA strand breaks and crosslinks induced by low energy electrons. Phys. Chem. Chem. Phys. 2016, 18, 32762–32771. [Google Scholar] [CrossRef]
- Brodeur, N.; Cloutier, P.; Bass, A.D.; Bertrand, G.; Hunting, D.J.; Grandbois, M.; Sanche, L. Absolute cross section for DNA damage induced by low-energy (10 eV) electrons: Experimental refinements and sample characterization by AFM. J. Chem. Phys. 2018, 149, 164904. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, W.; Brodeur, N.; Cloutier, P.; Zheng, Y.; Sanche, L. Absolute cross sections for chemoradiation therapy: Damages to cisplatin-DNA complexes induced by 10 eV electrons. J. Chem. Phys. 2019, 150, 195101. [Google Scholar] [CrossRef] [PubMed]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hunting, D.J.; Ayotte, P.; Sanche, L. Role of secondary electrons in the concomitant chemoradiation therapy of cancer. Phys. Rev. Lett. 2008, 100, 198101. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, M.; Hunting, D.J.; Sanche, L. New insights into the mechanism underlying the synergistic action of ionizing radiation with platinum chemotherapeutic drugs: The role of low-energy electrons. Int. J. Radiat. Oncol. 2013, 87, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, M.; Alizadeh, E.; Cloutier, P.; Hunting, D.J.; Sanche, L. A single subexcitation-energy electron can induce a double-strand break in DNA modified by platinum chemotherapeutic drugs. Chem. Med. Chem. 2014, 9, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Bao, Q.; Chen, Y.; Zheng, Y.; Sanche, L. Cisplatin radiosensitization of DNA irradiated with 2–20 eV electrons: Role of transient anions. J. Phys. Chem. C 2014, 118, 15516–15524. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhou, L.; Tian, Q.; Zheng, Y.; Sanche, L. Chemoradiation cancer therapy: Molecular mechanisms of cisplatin radiosensitization. J. Phys. Chem. C 2017, 121, 17505–17513. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, Y.; Zhou, L.; Shao, Y.; Fu, X.; Zheng, Y. Molecular efficacy of radio- and chemotherapy sequences from direct DNA damage measurements. Int. J. Radiat. Biol. 2017, 93, 1274–1282. [Google Scholar] [CrossRef]
- Li, Z.; Cloutier, P.; Sanche, L.; Wagner, J.R. Low energy electron induced damage in a trinucleotide containing 5-bromouracil. J. Phys. Chem. B 2011, 115, 13668–13673. [Google Scholar] [CrossRef]
- Simons, J. How do low-energy (0.1–2 eV) electrons cause DNA strand breaks? Acc. Chem. Res. 2006, 39, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zheng, Y.; Cloutier, P.; Sanche, L.; Wagner, J.R. Low energy electron induced DNA damage: Effects of terminal phosphate and base moieties on the distribution of damage. J. Am. Chem. Soc. 2008, 130, 5612–5613. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Poonia, V.S.; Fontanesi, C.; Naaman, R.; Fleming, A.M.; Burrows, C.J. The effect of oxidative damage on charge and spin transport in DNA. J. Am. Chem. Soc. 2019, 141, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Hoy, C.L.; Ferhanoglu, O.; Yildirim, M.; Kim, K.H.; Karajanagi, S.S.; Chan, K.M.C.; Kobler, J.B.; Zeitels, S.M.; Ben-Yakar, A. Clinical ultrafast laser surgery: Recent advances and future directions. IEEE J. Sel. Top. Quantum Electron. 2014, 20, 242–255. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Sanche, L. Clustered DNA Damages induced by 0.5 to 30 eV Electrons. Int. J. Mol. Sci. 2019, 20, 3749. https://doi.org/10.3390/ijms20153749
Zheng Y, Sanche L. Clustered DNA Damages induced by 0.5 to 30 eV Electrons. International Journal of Molecular Sciences. 2019; 20(15):3749. https://doi.org/10.3390/ijms20153749
Chicago/Turabian StyleZheng, Yi, and Léon Sanche. 2019. "Clustered DNA Damages induced by 0.5 to 30 eV Electrons" International Journal of Molecular Sciences 20, no. 15: 3749. https://doi.org/10.3390/ijms20153749