Overexpression of the Transcription Factors GmSHN1 and GmSHN9 Differentially Regulates Wax and Cutin Biosynthesis, Alters Cuticle Properties, and Changes Leaf Phenotypes in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Isolation and Identification of Soybean SHINE Homologs
2.2. GmSHN Genes Are Differentially Expressed in Various Organs of Soybean
2.3. Constitutive Expression of GmSHN Genes in Arabidopsis Results in Different Leaf Phenotypes
2.4. GmSHN1 and GmSHN9 Proteins Are Localized Primarily in the Nucleus
2.5. Overxpression of GmSHN1 and GmSHN9 Induces Epicuticular Wax Accumulation in Transgenic Arabidopsis Leaves
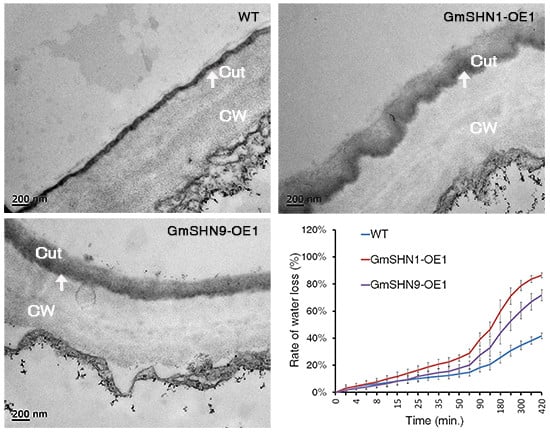
2.6. Overxpression of GmSHN1 and GmSHN9 Affects Cuticle Property and Alters Cuticle Composition in Transgenic Arabidopsis Leaves
2.7. Overxpression of GmSHN1 and GmSHN9 Induces Expression of Wax and Cutin Biosynthesis Related Genes in Transgenic Arabidopsis Plants
3. Discussion
3.1. GmSHN1 and GmSHN9 Differentially Regulate the Cuticular Wax Biosynthetic Pathway
3.2. GmSHN1 and GmSHN9 Are Involved in the Regulation of Cutin Biosynthesis, Especially Hydroxylation
3.3. GmSHN1 and GmSHN9 Are Involved in Regulation of Cuticle Property and Permeability
3.4. GmSHN1 and GmSHN9 Are Involved in Regulation of Leaf Development
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification and Analysis of GmSHN Genes
4.3. RNA Extraction and Semi-Quantitative RT-PCR
4.4. Plasmid Construction
4.5. Plant Transformation and Selection
4.6. Physiological Measurements
4.7. Quantitative Real-Time RT-PCR (qRT-PCR)
4.8. Protein Localization Analysis
4.9. Environmental Scanning Electron Microscopy Analysis
4.10. Wax Extraction and Chemical Analysis
4.11. Water Loss Analysis
4.12. Transmission Electron Microscope (TEM)
4.13. Cutin Extraction and Chemical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pollard, M.; Beisson, F.; Li, Y.; Ohlrogge, J.B. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008, 13, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed]
- Sieber, P.; Schorderet, M.; Ryser, U.; Buchala, A.; Kolattukudy, P.; Metraux, J.P.; Nawrath, C. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 2000, 12, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Xiong, L.M.; Li, W.B.; Zhu, J.K.; Zhu, J.H. The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell 2011, 23, 1971–1984. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Joubes, J. Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog. Lipid Res. 2013, 52, 110–129. [Google Scholar] [CrossRef] [PubMed]
- Riederer, M.; Schreiber, L. Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 2001, 52, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K. The formation and function of plant cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Kunst, L.; Samuels, A.L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003, 42, 51–80. [Google Scholar] [CrossRef]
- Suh, M.C.; Samuels, A.L.; Jetter, R.; Kunst, L.; Pollard, M.; Ohlrogge, J.; Beisson, F. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005, 139, 1649–1665. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdallat, A.M.; Al-Debei, H.S.; Ayad, J.Y.; Hasan, S. Over-expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in tomato. Int. J. Mol. Sci. 2014, 15, 19499–19515. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Peng, T.; Dai, W.S. Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 2014, 32, 303–317. [Google Scholar] [CrossRef]
- Go, Y.S.; Kim, H.; Kim, H.J.; Suh, M.C. Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 2014, 26, 1666–1680. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Broeckling, C.D.; Blancaflor, E.B.; Sledge, M.K.; Sumner, L.W.; Wang, Z.Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 2005, 42, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef] [PubMed]
- Broun, P.; Poindexter, P.; Osborne, E.; Jiang, C.Z.; Riechmann, J.L. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 4706–4711. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Wan, L.Y.; Zhang, L.X.; Zhang, Z.J.; Zhang, H.W.; Quan, R.D.; Zhou, S.R.; Huang, R.F. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012, 78, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.Y.; Jenks, M.A.; Liu, J.; Liu, A.L.; Zhang, X.W.; Xiang, J.H.; Zou, J.; Peng, Y.; Chen, X.B. Overexpression of transcription factor OsWR2 regulates wax and cutin biosynthesis in rice and enhances its tolerance to water deficit. Plant Mol. Biol. Rep. 2014, 32, 719–731. [Google Scholar] [CrossRef]
- Jager, K.; Misko, A.; Fabian, A.; Deak, C.; Kiss-Baba, E.; Polgari, D.; Barnabas, B.; Papp, I. Expression of a WIN/SHN-type regulator from wheat triggers disorganized proliferation in the Arabidopsis leaf cuticle. Biol. Plantarum. 2015, 59, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.X.; Adato, A.; Alkan, N.; He, Y.H.; Lashbrooke, J.; Matas, A.J.; Meir, S.; Malitsky, S.; Isaacson, T.; Prusky, D.; et al. The tomato SLSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol. 2013, 197, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Lashbrooke, J.; Aharoni, A.; Costa, F. Genome investigation suggests MdSHN3, an APETALA2-domain transcription factor gene, to be a positive regulator of apple fruit cuticle formation and an inhibitor of russet development. J. Exp. Bot. 2015, 66. [Google Scholar] [CrossRef] [PubMed]
- Kannangara, R.; Branigan, C.; Liu, Y.; Penfield, T.; Rao, V.; Mouille, G.; Hofte, H.; Pauly, M.; Riechmann, J.L.; Broun, P. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 2007, 19, 1278–1294. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.X.; Malitsky, S.; de Oliveira, S.; Branigan, C.; Franke, R.B.; Schreiber, L.; Aharoni, A. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 2011, 7, 1001388. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.; Buxdorf, K.; Shi, J.X.; Feldmesser, E.; Schreiber, L.; Aharoni, A.; Levy, M. Overexpression of AtSHN1/WIN1 provokes unique defense responses. PLoS ONE 2013, 8, 70146. [Google Scholar] [CrossRef] [PubMed]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Buggs, R.J.A. The consequences of polyploidy and hybridisation for transcriptome dynamics unravelling gene expression of complex crop genomes. Heredity 2013, 110, 97–98. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. The leaf index: Heteroblasty, natural variation, and the genetic control of polar processes of leaf expansion. Plant Cell Physiol. 2002, 43, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Kim, G.T.; Kim, I.J.; Park, J.; Kwak, S.S.; Choi, G.; Chung, W.I. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 2006, 133, 4305–4314. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. Leaf shape: Genetic controls and environmental factors. Int. J. Dev. Biol. 2005, 49, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mploc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, 11335. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Leide, J.; Hildebrandt, U.; Reussing, K.; Riederer, M.; Vogg, G. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: Effects of a deficiency in a β-ketoacyl-coenzyme a synthase (LeCER6). Plant Physiol. 2007, 144, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Cheng, Z.; Lei, C.; Xu, F.; Gao, S.; Ren, Y.; Wang, J.; Zhang, X.; Wu, F.; Guo, X.; et al. Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 2012, 235, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.; Post-Beittenmiller, D.; Jaworski, J.G. KCS1 encodes a fatty acid elongase 3-ketoacyl-coA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999, 17, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Jung, S.J.; Go, Y.S.; Kim, H.U.; Kim, J.K.; Cho, H.J.; Park, O.K.; Suh, M.C. Two Arabidopsis 3-ketoacyl coA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009, 60, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Mayfield, J.A.; Miley, N.L.; Chau, S.; Fischer, R.L.; Preuss, D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 2000, 12, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Lolle, S.J.; Berlyn, G.P.; Engstrom, E.M.; Krolikowski, K.A.; Reiter, W.D.; Pruitt, R.E. Developmental regulation of cell interactions in the Arabidopsis Fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev. Biol. 1997, 189, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, F.; Wu, X.Z.; Li, F.L.; Haslam, R.P.; Markham, J.E.; Zheng, H.Q.; Napier, J.A.; Kunst, L. Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 2009, 150, 1174–1191. [Google Scholar] [CrossRef] [PubMed]
- Bach, L.; Michaelson, L.V.; Haslam, R.; Bellec, Y.; Gissot, L.; Marion, J.; Da Costa, M.; Boutin, J.P.; Miquel, M.; Tellier, F.; et al. The very-long-chain hydroxy fatty acyl-coA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 2008, 105, 14727–14731. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.Q.; Rowland, O.; Kunst, L. Disruptions of the Arabidopsis enoyl-coA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 2005, 17, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Haslam, T.M.; Manas-Fernandez, A.; Zhao, L.F.; Kunst, L. Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 2012, 160, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubes, J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Lee, R.; Franke, R.; Schreiber, L.; Kunst, L. The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 2007, 581, 3538–3544. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.; Zhao, L.F.; McFarlane, H.E.; Aiga, M.; Lam, V.; Hooker, T.S.; Kunst, L. RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 2012, 159, 1385–1395. [Google Scholar] [CrossRef] [PubMed]
- Bourdenx, B.; Bernard, A.; Domergue, F.; Pascal, S.; Leger, A.; Roby, D.; Pervent, M.; Vile, D.; Haslam, R.P.; Napier, J.A.; et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011, 156, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme a reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Bonaventure, G.; Beisson, F.; Ohlrogge, J.; Pollard, M. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 2004, 40, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Franke, R.; Briesen, I.; Wojciechowski, T.; Faust, A.; Yephremov, A.; Nawrath, C.; Schreiber, L. Apoplastic polyesters in Arabidopsis surface tissues—A typical suberin and a particular cutin. Phytochemistry 2005, 66, 2643–2658. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Pollard, M.; Sauveplane, V.; Pinot, F.; Ohlrogge, J.; Beisson, F. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. USA 2009, 106, 22008–22013. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Yu, K.S.; Gao, Q.M.; Wilson, E.V.; Navarre, D.; Kachroo, P.; Kachroo, A. Acyl coA binding proteins are required for cuticle formation and plant responses to microbes. Front. Plant Sci. 2012, 3, 224. [Google Scholar] [CrossRef] [PubMed]
- Menard, R.; Verdier, G.; Ors, M.; Erhardt, M.; Beisson, F.; Shen, W.H. Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Wellesen, K.; Durst, F.; Pinot, F.; Benveniste, I.; Nettesheim, K.; Wisman, E.; Steiner-Lange, S.; Saedler, H.; Yephremov, A. Functional analysis of the lacerata gene of Arabidopsis provides evidence for different robes of fatty acid omega-hydroxylation in development. Proc. Natl. Acad. Sci. USA 2001, 98, 9694–9699. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.Y.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009, 59, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Pulsifer, I.P.; Kluge, S.; Rowland, O. Arabidopsis LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1), LACS2, and LACS3 facilitate fatty acid uptake in yeast. Plant Physiol. Biochem. 2012, 51, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Schnurr, J.; Shockey, J.; Browse, J. The acyl-coA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 2004, 16, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Bird, D.; Beisson, F.; Brigham, A.; Shin, J.; Greer, S.; Jetter, R.; Kunst, L.; Wu, X.W.; Yephremov, A.; Samuels, L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 2007, 52, 485–498. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.E.; Shin, J.J.H.; Bird, D.A.; Samuels, A.L. Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 2010, 22, 3066–3075. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Goodwin, S.M.; Boroff, V.L.; Liu, X.; Jenks, M.A. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 2003, 15, 1170–1185. [Google Scholar] [CrossRef] [PubMed]
- Park, J.J.; Jin, P.; Yoon, J.; Yang, J.I.; Jeong, H.J.; Ranathunge, K.; Schreiber, L.; Franke, R.; Lee, I.J.; An, G. Mutation in Wilted Dwarf and Lethal 1 (WDL1) causes abnormal cuticle formation and rapid water loss in rice. Plant Mol. Biol. 2010, 74, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Li, M.C.; Zhao, M.M.; Wu, H.Y.; Wu, W.; Xu, Y.N. Cloning, characterization and functional analysis of two type 1 diacylglycerol acyltransferases (DGAT1s) from tetraena mongolica. J. Integr. Plant Biol. 2013, 55, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Xu, Y.N. Hypocotyl-based agrobacterium-mediated transformation of soybean (Glycine max) and application for RNA interference. Plant Cell Rep. 2008, 27, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.L.; Liu, H.J.; Wang, X.R.; Zeng, Q.Y. Molecular evolution and expression divergence of the populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytologist 2013, 197, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids—Pigments of photosynthetic biomembranes. Method Enzymol. 1987, 148, 350–382. [Google Scholar]
- Zhao, S.Z.; Luo, Y.Z.; Zhang, Z.L.; Xu, M.Y.; Wang, W.B.; Zhao, Y.M.; Zhang, L.; Fan, Y.L.; Wang, L. ZmSOC1, an MADS-box transcription factor from Zea mays, promotes flowering in Arabidopsis. Int. J. Mol. Sci. 2014, 15, 19987–20003. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Liu, C.; Li, M.C.; Zhao, M.M.; Gu, D.; Xu, Y.N. Effects of monogalactoglycerolipid deficiency and diacylglycerol acyltransferase overexpression on oil accumulation in transgenic tobacco. Plant Mol. Biol. Rep. 2013, 31, 1077–1088. [Google Scholar] [CrossRef]
- Wang, L.Y.; Ouyang, M.; Li, Q.N.; Zou, M.J.; Guo, J.K.; Ma, J.F.; Lu, C.M.; Zhang, L.X. The Arabidopsis chloroplast ribosome recycling factor is essential for embryogenesis and chloroplast biogenesis. Plant Mol. Biol. 2010, 74, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Damm, B.; Schmidt, R.; Willmitzer, L. Efficient transformation of Arabidopsis thaliana using direct gene-transfer to protoplasts. Mol. Gen. Genet. 1989, 217, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, F.; Ebert, M.; Worringen, A.; Schutz, L.; Weinbruch, S. Environmental scanning electron microscopy (ESEM) as a new technique to determine the ice nucleation capability of individual atmospheric aerosol particles. Atmos. Environ. 2007, 41, 8219–8227. [Google Scholar] [CrossRef]
- Kosma, D.K.; Bourdenx, B.; Bernard, A.; Parsons, E.P.; Lu, S.; Joubes, J.; Jenks, M.A. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009, 151, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Domergue, F.; Vishwanath, S.J.; Joubes, J.; Ono, J.; Lee, J.A.; Bourdon, M.; Alhattab, R.; Lowe, C.; Pascal, S.; Lessire, R.; et al. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 2010, 153, 1539–1554. [Google Scholar] [CrossRef] [PubMed]











© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Wu, H.; Zhao, M.; Wu, W.; Xu, Y.; Gu, D. Overexpression of the Transcription Factors GmSHN1 and GmSHN9 Differentially Regulates Wax and Cutin Biosynthesis, Alters Cuticle Properties, and Changes Leaf Phenotypes in Arabidopsis. Int. J. Mol. Sci. 2016, 17, 587. https://doi.org/10.3390/ijms17040587
Xu Y, Wu H, Zhao M, Wu W, Xu Y, Gu D. Overexpression of the Transcription Factors GmSHN1 and GmSHN9 Differentially Regulates Wax and Cutin Biosynthesis, Alters Cuticle Properties, and Changes Leaf Phenotypes in Arabidopsis. International Journal of Molecular Sciences. 2016; 17(4):587. https://doi.org/10.3390/ijms17040587
Chicago/Turabian StyleXu, Yangyang, Hanying Wu, Mingming Zhao, Wang Wu, Yinong Xu, and Dan Gu. 2016. "Overexpression of the Transcription Factors GmSHN1 and GmSHN9 Differentially Regulates Wax and Cutin Biosynthesis, Alters Cuticle Properties, and Changes Leaf Phenotypes in Arabidopsis" International Journal of Molecular Sciences 17, no. 4: 587. https://doi.org/10.3390/ijms17040587