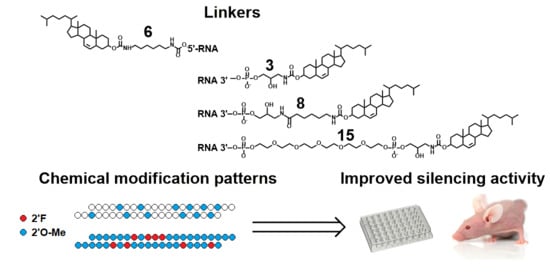
Cholesterol Conjugates of Small Interfering RNA: Linkers and Patterns of Modification
Abstract
:1. Introduction
2. Results
2.1. Silencing Activity of Cholesterol-Modified siRNA under Transfection
2.2. Silencing Activity of Cholesterol-Modified siRNA in Carrier-Free Mode
2.3. Silencing Activity of Selectively and Fully Modified siRNA
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. Cell Cultures
4.3. Silencing Activity Assay Using Flow Cytometry
4.4. Mice
4.5. Silencing Activity Assay in KB-8-5 Xenograft Tumors in SCID Mice after Intravenous Administration
- hMDR1 forward: 5′-CCGATACATGGTTTTCCGATCC-3′,
- hMDR1 reverse: 5′-CAGCAAGCCTGGAACCTATAG-3′,
- hMDR1 probe: ((5,6)-FAM)-5′-AACTTGAGCAGCATCATTGGCGAG-3′-BHQ1,
- hHPRT forward: 5′-TGCTGAGGATTTGGAAAGGG-3′
- hHPRT reverse: 5′-ACAGAGGGCTACAATGTGATG-3′,
- hHPRT probe: ((5,6)-Rox)-5′-AGGACTGAACGTCTTGCTCGAGATG-3′-BHQ2,
4.6. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The Current Landscape of Nucleic Acid Therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic SiRNA: State of the Art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Corydon, I.J.; Fabian-Jessing, B.K.; Jakobsen, T.S.; Jørgensen, A.C.; Jensen, E.G.; Askou, A.L.; Aagaard, L.; Corydon, T.J. 25 Years of Maturation: A Systematic Review of RNAi in the Clinic. Mol. Ther.—Nucleic Acids 2023, 33, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Manoharan, M. Chemistry, Structure and Function of Approved Oligonucleotide Therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef]
- Liu, C.; Shi, Q.; Huang, X.; Koo, S.; Kong, N.; Tao, W. MRNA-Based Cancer Therapeutics. Nat. Rev. Cancer 2023, 23, 526–543. [Google Scholar] [CrossRef]
- Chernikov, I.V.; Vlassov, V.V.; Chernolovskaya, E.L. Current Development of SiRNA Bioconjugates: From Research to the Clinic. Front. Pharmacol. 2019, 10, 444. [Google Scholar] [CrossRef]
- Salim, L.; Islam, G.; Desaulniers, J.P. Targeted Delivery and Enhanced Gene-Silencing Activity of Centrally Modified Folic Acid-SiRNA Conjugates. Nucleic Acids Res. 2020, 48, 75–85. [Google Scholar] [CrossRef]
- Nair, J.K.; Willoughby, J.L.S.; Chan, A.; Charisse, K.; Alam, M.R.; Wang, Q.; Hoekstra, M.; Kandasamy, P.; Kelin, A.V.; Milstein, S.; et al. Multivalent N-Acetylgalactosamine-Conjugated SiRNA Localizes in Hepatocytes and Elicits Robust RNAi-Mediated Gene Silencing. J. Am. Chem. Soc. 2014, 136, 16958–16961. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Holland, R.; Wood, M.; Pasetka, C.; Palmer, L.; Samaridou, E.; McClintock, K.; Borisevich, V.; Geisbert, T.W.; Cross, R.W.; et al. Combination Treatment of Mannose and GalNAc Conjugated Small Interfering RNA Protects against Lethal Marburg Virus Infection. Mol. Ther. 2023, 31, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, F.; Xu, Y.; Zhang, W.; Hu, Y.; Fu, Y.; Xu, W.; Ge, S.; Fan, X.; Lu, L. Cholesterol Modification of SDF-1-Specific SiRNA Enables Therapeutic Targeting of Angiogenesis through Akt Pathway Inhibition. Exp. Eye Res. 2019, 184, 64–71. [Google Scholar] [CrossRef]
- Zhou, J.; Lazar, D.; Li, H.; Xia, X.; Satheesan, S.; Charlins, P.; O’Mealy, D.; Akkina, R.; Saayman, S.; Weinberg, M.S.; et al. Receptor-Targeted Aptamer-SiRNA Conjugate-Directed Transcriptional Regulation of HIV-1. Theranostics 2018, 8, 1575–1590. [Google Scholar] [CrossRef] [PubMed]
- Cen, B.; Wei, Y.; Huang, W.; Teng, M.; He, S.; Li, J.; Wang, W.; He, G.; Bai, X.; Liu, X.; et al. An Efficient Bivalent Cyclic RGD-PIK3CB SiRNA Conjugate for Specific Targeted Therapy against Glioblastoma In Vitro and In Vivo. Mol. Ther.—Nucleic Acids 2018, 13, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Smidt, J.M.; Lykke, L.; Stidsen, C.E.; Pristovšek, N.; Gothelf, K.V. Synthesis of Peptide–SiRNA Conjugates via Internal Sulfonylphosphoramidate Modifications and Evaluation of Their in Vitro Activity. Nucleic Acids Res. 2023, 52, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Malecova, B.; Burke, R.S.; Cochran, M.; Hood, M.D.; Johns, R.; Kovach, P.R.; Doppalapudi, V.R.; Erdogan, G.; Arias, J.D.; Darimont, B.; et al. Targeted Tissue Delivery of RNA Therapeutics Using Antibody-Oligonucleotide Conjugates (AOCs). Nucleic Acids Res. 2023, 51, 5901–5910. [Google Scholar] [CrossRef]
- Brown, K.M.; Nair, J.K.; Janas, M.M.; Anglero-Rodriguez, Y.I.; Dang, L.T.H.; Peng, H.; Theile, C.S.; Castellanos-Rizaldos, E.; Brown, C.; Foster, D.; et al. Expanding RNAi Therapeutics to Extrahepatic Tissues with Lipophilic Conjugates. Nat. Biotechnol. 2022, 40, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Benizri, S.; Gissot, A.; Martin, A.; Vialet, B.; Grinstaff, M.W.; Barthélémy, P. Bioconjugated Oligonucleotides: Recent Developments and Therapeutic Applications. Bioconjug. Chem. 2019, 30, 366–383. [Google Scholar] [CrossRef] [PubMed]
- Chernikov, I.V.; Gladkikh, D.V.; Meschaninova, M.I.; Ven’yaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Cholesterol-Containing Nuclease-Resistant SiRNA Accumulates in Tumors in a Carrier-Free Mode and Silences MDR1 Gene. Mol. Ther.—Nucleic Acids 2017, 6, 209–220. [Google Scholar] [CrossRef]
- Engelbeen, S.; Pasteuning-Vuhman, S.; Boertje-Van Der Meulen, J.; Parmar, R.; Charisse, K.; Sepp-Lorenzino, L.; Manoharan, M.; Aartsma-Rus, A.; Van Putten, M. Efficient Downregulation of Alk4 in Skeletal Muscle After Systemic Treatment with Conjugated SiRNAs in a Mouse Model for Duchenne Muscular Dystrophy. Nucleic Acid Ther. 2023, 33, 26–34. [Google Scholar] [CrossRef]
- Davis, S.M.; Hariharan, V.N.; Lo, A.; Turanov, A.A.; Echeverria, D.; Sousa, J.; McHugh, N.; Biscans, A.; Alterman, J.F.; Karumanchi, S.A.; et al. Chemical Optimization of SiRNA for Safe and Efficient Silencing of Placental SFLT1. Mol. Ther.—Nucleic Acids 2022, 29, 135–149. [Google Scholar] [CrossRef]
- Biscans, A.; Coles, A.; Haraszti, R.; Echeverria, D.; Hassler, M.; Osborn, M.; Khvorova, A. Diverse Lipid Conjugates for Functional Extra-Hepatic SiRNA Delivery In Vivo. Nucleic Acids Res. 2019, 47, 1082–1096. [Google Scholar] [CrossRef]
- Turanov, A.A.; Lo, A.; Hassler, M.R.; Makris, A.; Ashar-Patel, A.; Alterman, J.F.; Coles, A.H.; Haraszti, R.A.; Roux, L.; Godinho, B.M.D.C.; et al. RNAi Modulation of Placental SFLT1 for the Treatment of Preeclampsia. Nat. Biotechnol. 2018, 36, 1164–1173. [Google Scholar] [CrossRef]
- Tang, Q.; Sousa, J.; Echeverria, D.; Fan, X.; Hsueh, Y.C.; Afshari, K.; MeHugh, N.; Cooper, D.A.; Vangjeli, L.; Monopoli, K.; et al. RNAi-Based Modulation of IFN-γ Signaling in Skin. Mol. Ther. 2022, 30, 2709–2721. [Google Scholar] [CrossRef]
- Tang, Q.; Gross, K.Y.; Fakih, H.H.; Jackson, S.O.; Zain, U.I.; Abideen, M.; Monopoli, K.R.; Blanchard, C.; Bouix-Peter, C.; Portal, T.; et al. Multispecies-Targeting SiRNAs for the Modulation of JAK1 in the Skin. Mol. Ther. Nucleic Acids 2024, 35, 102117. [Google Scholar] [CrossRef] [PubMed]
- Witzigmann, D.; Kulkarni, J.A.; Leung, J.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Therapeutic Gene Regulation in the Liver. Adv. Drug Deliv. Rev. 2020, 159, 344–363. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Schlegel, M.K.; Manoharan, M. Acyclic (S)-Glycol Nucleic Acid (S-GNA) Modification of SiRNAs Improves the Safety of RNAi Therapeutics While Maintaining Potency. RNA 2023, 29, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Bala, S.; Liao, J.Y.; Datta, D.; Mikami, A.; Woods, L.; Harp, J.M.; Gilbert, J.A.; Bisbe, A.; Manoharan, R.M.; et al. Shorter Is Better: The α-(l)-Threofuranosyl Nucleic Acid Modification Improves Stability, Potency, Safety, and Ago2 Binding and Mitigates Off-Target Effects of Small Interfering RNAs. J. Am. Chem. Soc. 2023, 145, 19691–19706. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Iwamoto, N.; Marappan, S.; Luu, K.; Tripathi, S.; Purcell-Estabrook, E.; Shelke, J.D.; Shah, H.; Lamattina, A.; Pan, Q.; et al. Impact of Stereopure Chimeric Backbone Chemistries on the Potency and Durability of Gene Silencing by RNA Interference. Nucleic Acids Res. 2023, 51, 4126–4147. [Google Scholar] [CrossRef] [PubMed]
- Volkov, A.A.; Kruglova, N.S.; Meschaninova, M.I.; Venyaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Selective Protection of Nuclease-Sensitive Sites in SiRNA Prolongs Silencing Effect. Oligonucleotides 2009, 19, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.J.; Brown, C.R.; Shaikh, S.; Trapp, C.; Schlegel, M.K.; Qian, K.; Sehgal, A.; Rajeev, K.G.; Jadhav, V.; Manoharan, M.; et al. Advanced SiRNA Designs Further Improve In Vivo Performance of GalNAc-SiRNA Conjugates. Mol. Ther. 2018, 26, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Petrova, N.S.; Chernikov, I.V.; Meschaninova, M.I.; Dovydenko, I.S.; Venyaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Carrier-Free Cellular Uptake and the Gene-Silencing Activity of the Lipophilic SiRNAs Is Strongly Affected by the Length of the Linker between SiRNA and Lipophilic Group. Nucleic Acids Res. 2012, 40, 2330–2344. [Google Scholar] [CrossRef] [PubMed]
- Chernikov, I.V.; Gladkikh, D.V.; Meschaninova, M.I.; Karelina, U.A.; Ven’Yaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Fluorophore Labeling Affects the Cellular Accumulation and Gene Silencing Activity of Cholesterol-Modified SiRNAs In Vitro. Nucleic Acid Ther. 2019, 29, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.F.; Coles, A.H.; Biscans, A.; Haraszti, R.A.; Roux, L.; Davis, S.; Ly, S.; Echeverria, D.; Hassler, M.R.; Godinho, B.M.D.C.; et al. Hydrophobicity Drives the Systemic Distribution of Lipid-Conjugated SiRNAs via Lipid Transport Pathways. Nucleic Acids Res. 2019, 47, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Janas, M.M.; Schlegel, M.K.; Harbison, C.E.; Yilmaz, V.O.; Jiang, Y.; Parmar, R.; Zlatev, I.; Castoreno, A.; Xu, H.; Shulga-Morskaya, S.; et al. Selection of GalNAc-Conjugated SiRNAs with Limited off-Target-Driven Rat Hepatotoxicity. Nat. Commun. 2018, 9, 723. [Google Scholar] [CrossRef] [PubMed]
- Biscans, A.; Caiazzi, J.; Davis, S.; McHugh, N.; Sousa, J.; Khvorova, A. The Chemical Structure and Phosphorothioate Content of Hydrophobically Modified SiRNAs Impact Extrahepatic Distribution and Efficacy. Nucleic Acids Res. 2020, 48, 7665–7680. [Google Scholar] [CrossRef] [PubMed]
- Godinho, B.M.D.C.; Gilbert, J.W.; Haraszti, R.A.; Coles, A.H.; Biscans, A.; Roux, L.; Nikan, M.; Echeverria, D.; Hassler, M.; Khvorova, A. Pharmacokinetic Profiling of Conjugated Therapeutic Oligonucleotides: A High-Throughput Method Based Upon Serial Blood Microsampling Coupled to Peptide Nucleic Acid Hybridization Assay. Nucleic Acid Ther. 2017, 27, 323–334. [Google Scholar] [CrossRef]
- Wolfrum, C.; Shi, S.; Jayaprakash, K.N.; Jayaraman, M.; Wang, G.; Pandey, R.K.; Rajeev, K.G.; Nakayama, T.; Charrise, K.; Ndungo, E.M.; et al. Mechanisms and Optimization of In Vivo Delivery of Lipophilic SiRNAs. Nat. Biotechnol. 2007, 25, 1149–1157. [Google Scholar] [CrossRef]
- Ly, S.; Navaroli, D.M.; Didiot, M.-C.; Cardia, J.; Pandarinathan, L.; Alterman, J.F.; Fogarty, K.; Standley, C.; Lifshitz, L.M.; Bellve, K.D.; et al. Visualization of Self-Delivering Hydrophobically Modified SiRNA Cellular Internalization. Nucleic Acids Res. 2017, 45, 15–25. [Google Scholar] [CrossRef]
- Alterman, J.F.; Hall, L.M.; Coles, A.H.; Hassler, M.R.; Didiot, M.-C.; Chase, K.; Abraham, J.; Sottosanti, E.; Johnson, E.; Sapp, E.; et al. Hydrophobically Modified SiRNAs Silence Huntingtin MRNA in Primary Neurons and Mouse Brain. Mol. Ther.—Nucleic Acids 2015, 4, e266. [Google Scholar] [CrossRef]
- Wada, S.; Yasuhara, H.; Wada, F.; Sawamura, M.; Waki, R.; Yamamoto, T.; Harada-Shiba, M.; Obika, S. Evaluation of the Effects of Chemically Different Linkers on Hepatic Accumulations, Cell Tropism and Gene Silencing Ability of Cholesterol-Conjugated Antisense Oligonucleotides. J. Control. Release 2016, 226, 57–65. [Google Scholar] [CrossRef]
- Hassler, M.R.; Turanov, A.A.; Alterman, J.F.; Haraszti, R.A.; Coles, A.H.; Osborn, M.F.; Echeverria, D.; Nikan, M.; Salomon, W.E.; Roux, L.; et al. Comparison of Partially and Fully Chemically-Modified SiRNA in Conjugate-Mediated Delivery In Vivo. Nucleic Acids Res. 2018, 46, 2185–2196. [Google Scholar] [CrossRef] [PubMed]
- Nair, J.K.; Attarwala, H.; Sehgal, A.; Wang, Q.; Aluri, K.; Zhang, X.; Gao, M.; Liu, J.; Indrakanti, R.; Schofield, S.; et al. Impact of Enhanced Metabolic Stability on Pharmacokinetics and Pharmacodynamics of GalNAc-SiRNA Conjugates. Nucleic Acids Res. 2017, 45, 10969–10977. [Google Scholar] [CrossRef] [PubMed]
- Laurent, Q.; Martinent, R.; Moreau, D.; Winssinger, N.; Sakai, N.; Matile, S. Oligonucleotide Phosphorothioates Enter Cells by Thiol-Mediated Uptake. Angew. Chem.—Int. Ed. 2021, 60, 19102–19106. [Google Scholar] [CrossRef] [PubMed]
- Shmushkovich, T.; Monopoli, K.R.; Homsy, D.; Leyfer, D.; Betancur-Boissel, M.; Khvorova, A.; Wolfson, A.D. Functional Features Defining the Efficacy of Cholesterol-Conjugated, Self-Deliverable, Chemically Modified SiRNAs. Nucleic Acids Res. 2018, 46, 10905–10916. [Google Scholar] [CrossRef]
- Zenkov, A.N.; Scvortsova, N.V.; Chernolovskaya, E.L.; Pospelova, T.I.; Vlassov, V.V. Expression of the MDR1 and MRP Genes in Patients with Lymphoma with Primary Bone Marrow Involvement. Nucleosides Nucleotides Nucleic Acids 2004, 23, 843–847. [Google Scholar] [CrossRef]
- Meschaninova, M.I.; Novopashina, D.S.; Semikolenova, O.A.; Silnikov, V.N.; Venyaminova, A.G. Novel Convenient Approach to the Solid-Phase Synthesis of Oligonucleotide Conjugates. Molecules 2019, 24, 4266. [Google Scholar] [CrossRef]
- Evdokimov, A.; Petruseva, I.; Tsidulko, A.; Koroleva, L.; Serpokrylova, I.; Silnikov, V.; Lavrik, O. New Synthetic Substrates of Mammalian Nucleotide Excision Repair System. Nucleic Acids Res. 2013, 41, e123. [Google Scholar] [CrossRef]



| Designation 1 | Sequence 5′-3′ 2 |
|---|---|
| MDR1 S | GGCUUmGACmAAGUUmGUmAUmAUmGG |
| Ch(6)-MDR1 S | Ch(6)-GGCUUmGACmAAGUUmGUmAUmAUmGG |
| MDR1-(n)Ch S, n = 3, 8, 15 | GGCUUmGACmAAGUUmGUmAUmAUmGG-(n)Ch |
| MDR1_−2G-(n)Ch S, n = 3, 8, 15 | GGCUUmGACmAAGUUmGUmAUmAUm-(n)Ch |
| MDR1_FM S | GmGmCmUmUmGmAfCmAfAfGfUmUmGmUmAmUmAmUmGmGm |
| MDR1 AS | AUmAUmACmAACUUmGUCmAAGCCmAA |
| MDR1_FM AS | AmUfAmUmAmCfAmAmCmUmUmGmUmCfAmAfGmCmCmAmAm |
| SCRm AS | CmAAGUCUCGUmAUmGUmAGUmGGUU |
| SCRm S SCR_FM_AS Ch(6)-SCR_FM S | CCmACUmACmAUmACGAGACUUmGUU CmAfAmGmUmCfUmCmGmUmAmUmGmUfAmGfUmGmGmUmUm Ch-CmCmAmCmUmAmCfAmUfAfCfGmAmGmAmCmUmUmGmUmUm |
| Designation | IC50, nM 1 |
|---|---|
| siMDR1 | 4.4 ± 1.9 # |
| siMDR1_AS_FM | 1.2 ± 0.3 ##,** |
| siMDR1_FM | 0.5 ± 0.3 ##,** |
| Ch(6)-siMDR1 | 15.3 ± 5.7 * |
| Ch(6)-siMDR1_AS_FM | 3.3 ± 2.2 # |
| Ch(6)-siMDR1_FM | 0.3 ± 0.1 ##,** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernikov, I.V.; Ponomareva, U.A.; Meschaninova, M.I.; Bachkova, I.K.; Vlassov, V.V.; Zenkova, M.A.; Chernolovskaya, E.L. Cholesterol Conjugates of Small Interfering RNA: Linkers and Patterns of Modification. Molecules 2024, 29, 786. https://doi.org/10.3390/molecules29040786
Chernikov IV, Ponomareva UA, Meschaninova MI, Bachkova IK, Vlassov VV, Zenkova MA, Chernolovskaya EL. Cholesterol Conjugates of Small Interfering RNA: Linkers and Patterns of Modification. Molecules. 2024; 29(4):786. https://doi.org/10.3390/molecules29040786
Chicago/Turabian StyleChernikov, Ivan V., Ul’yana A. Ponomareva, Mariya I. Meschaninova, Irina K. Bachkova, Valentin V. Vlassov, Marina A. Zenkova, and Elena L. Chernolovskaya. 2024. "Cholesterol Conjugates of Small Interfering RNA: Linkers and Patterns of Modification" Molecules 29, no. 4: 786. https://doi.org/10.3390/molecules29040786