Bacillus cereus Toxin Repertoire: Diversity of (Iso)cereulide(s)
Abstract
:1. Introduction
2. Results
3. Discussion
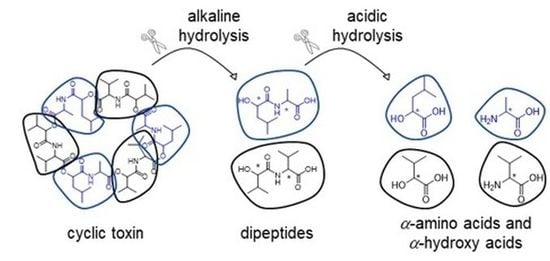
4. Experimental Section
General Experimental Procedures




Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ehling-Schulz, M.; Lereclus, D.; Koehler, T.M. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol Spectr. 2019, 7, 2018. [Google Scholar] [CrossRef] [PubMed]
- Makarasen, A.; Yoza, K.; Isobe, M. Higher Structure of Cereulide, an Emetic Toxin from Bacillus cereus, and Special Comparison with Valinomycin, an Antibiotic from Streptomyces fulvissimus. Chem. Asian J. 2009, 4, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Agata, N.; Mori, M.; Ohta, M.; Suwan, S.; Ohtani, I.; Isobe, M. A novel dodecadepsipeptide, cereulide, isolated from Bacillus cereus causes vacuole formation in HEp-2 cells. FEMS Microbiol. Lett. 1994, 121, 31–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suwan, S.; Isobe, M.; Ohtani, I.; Agata, N.; Mori, M.; Ohta, M.J. Structure of cereulide, a cyclic dodecadepsipeptide toxin from Bacillus cereus and studies on NMR characteristics of its alkali metal complexes including a conformational structure of the K+ complex. J. Chem. Soc., Perkin Trans. 1 1995, 1, 765–775. [Google Scholar] [CrossRef]
- Heberlig, G.W.; Boddy, C.N. Thioesterase from Cereulide Biosynthesis Is Responsible for Oligomerization and Macrocyclization of a Linear Tetradepsipeptide. J. Nat. Prod. 2020, 83, 1990–1997. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Fricker, M.; Scherer, S. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 2004, 48, 479–487. [Google Scholar] [CrossRef]
- Stenfors Arnesen, L.P.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef] [Green Version]
- Rouzeau-Szynalski, K.; Stollewerk, K.; Messelhäusser, U.; Ehling-Schulz, M. Why be serious about emetic Bacillus cereus: Cereulide production and industrial challenges. Food Microbiol. 2020, 85, 103279. [Google Scholar] [CrossRef]
- Kranzler, M.; Stollewerk, K.; Rouzeau-Szynalski, K.; Blayo, L.; Sulyok, M.; Ehling-Schulz, M. Temperature Exerts Control of Bacillus cereus Emetic Toxin Production on Post-transcriptional Levels. Front. Microbiol. 2016, 7, 1640. [Google Scholar] [CrossRef] [Green Version]
- Pitchayawasin, S.; Isobe, M.; Kuse, M.; Franz, T.; Agata, N.; Ohta, M. Molecular diversity of cereulide detected by means of nano-HPLC-ESI-Q-TOF-MS. Int. J. Mass Spectrom. 2004, 235, 123–129. [Google Scholar] [CrossRef]
- Stark, T.; Marxen, S.; Rütschle, A.; Lücking, G.; Scherer, S.; Ehling-Schulz, M.; Hofmann, T. Mass spectrometric profiling of Bacillus cereus strains and quantitation of the emetic toxin cereulide by means of stable isotope dilution analysis and HEp-2 bioassay. Anal. Bioanal. Chem. 2013, 405, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Marxen, S.; Stark, T.D.; Frenzel, E.; Rütschle, A.; Lücking, G.; Pürstinger, G.; Pohl, E.E.; Scherer, S.; Ehling-Schulz, M.; Hofmann, T. Chemodiversity of cereulide, the emetic toxin of Bacillus cereus. Anal. Bioanal. Chem. 2015, 407, 2439–2453. [Google Scholar] [CrossRef] [PubMed]
- Walser, V.; Kranzler, M.; Ehling-Schulz, M.; Stark, T.D.; Hofmann, T.F. Structure Revision of Isocereulide A, an Isoform of the Food Poisoning Emetic Bacillus cereus Toxin Cereulide. Molecules 2021, 26, 1360. [Google Scholar] [CrossRef] [PubMed]
- Naka, T.; Hattori, Y.; Takenaka, H.; Ohta, Y.; Kirihata, M.; Tanimori, S. Synthesis of the reported structure of homocereulide and its vacuolation assay. Bioorganic Med. Chem. Lett. 2019, 29, 734–739. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Vukov, N.; Schulz, A.; Shaheen, R.; Andersson, M.; Märtlbauer, E.; Scherer, S. Identification and Partial Characterization of the Nonribosomal Peptide Synthetase Gene Responsible for Cereulide Production in Emetic Bacillus cereus. Appl. Environ. Microbiol. 2005, 71, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Ehling-Schulz, M.; Fricker, M.; Grallert, H.; Rieck, P.; Wagner, M.; Scherer, S. Cereulide synthetase gene cluster from emetic Bacillus cereus: Structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 2006, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Rasko, D.A.; Rosovitz, M.J.; Økstad, O.A.; Fouts, D.E.; Jiang, L.; Cer, R.Z.; Kolstø, A.-B.; Gill, S.R.; Ravel, J. Complete Sequence Analysis of Novel Plasmids from Emetic and Periodontal Bacillus cereus Isolates Reveals a Common Evolutionary History among the B. cereus-Group Plasmids, Including Bacillus anthracis pXO1. J. Bacteriol. 2007, 189, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Dommel, M.K.; Frenzel, E.; Strasser, B.; Blöchinger, C.; Scherer, S.; Ehling-Schulz, M. Identification of the Main Promoter Directing Cereulide Biosynthesis in Emetic Bacillus cereus and Its Application for Real-Time Monitoring of ces Gene Expression in Foods. Appl. Environ. Microbiol. 2010, 76, 1232–1240. [Google Scholar] [CrossRef] [Green Version]
- Lücking, G.; Frenzel, E.; Rütschle, A.; Marxen, S.; Stark, T.D.; Hofmann, T.; Scherer, S.; Ehling-Schulz, M. Ces locus embedded proteins control the non-ribosomal synthesis of the cereulide toxin in emetic Bacillus cereus on multiple levels. Front. Microbiol. 2015, 6, 1101. [Google Scholar] [CrossRef] [Green Version]
- Gacek-Matthews, A.; Chromiková, Z.; Sulyok, M.; Lücking, G.; Barák, I.; Ehling-Schulz, M. Beyond Toxin Transport: Novel Role of ABC Transporter for Enzymatic Machinery of Cereulide NRPS Assembly Line. mBio 2020, 11, e01577-20. [Google Scholar] [CrossRef]
- Dietrich, R.; Jessberger, N.; Ehling-Schulz, M.; Märtlbauer, E.; Granum, P.E. The Food Poisoning Toxins of Bacillus cereus. Toxins 2021, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Magarvey, N.A.; Ehling-Schulz, M.; Walsh, C.T. Characterization of the Cereulide NRPS α-Hydroxy Acid Specifying Modules: Activation of α-Keto Acids and Chiral Reduction on the Assembly Line. J. Am. Chem. Soc. 2006, 128, 10698–10699. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, D.A.; Magarvey, N.A.; Schmeing, T.M. Characterization of Cereulide Synthetase, a Toxin-Producing Macromolecular Machine. PLoS ONE 2015, 10, e0128569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marxen, S.; Stark, T.D.; Rütschle, A.; Lücking, G.; Frenzel, E.; Ehling-Schulz, M.; Scherer, S.; Hofmann, T. Depsipeptide Intermediates Interrogate Proposed Biosynthesis of Cereulide, the Emetic Toxin of Bacillus cereus. Sci. Rep. 2015, 5, 10637. [Google Scholar] [CrossRef] [Green Version]
- Huguenin-Dezot, N.; Alonzo, D.A.; Heberlig, G.W.; Mahesh, M.; Nguyen, D.P.; Dornan, M.H.; Boddy, C.N.; Schmeing, T.M.; Chin, J.W. Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid. Nature 2019, 565, 112–117. [Google Scholar] [CrossRef]
- Pitchayawasin, S.; Kuse, M.; Koga, K.; Isobe, M.; Agata, N.; Ohta, M. Complexation of cyclic dodecadepsipeptide, Cereulide with ammonium salts. Bioorganic Med. Chem. Lett. 2003, 13, 3507–3512. [Google Scholar] [CrossRef]
- Mikkola, R.; Saris, N.-E.L.; Grigoriev, P.A.; Andersson Maria, A.; Salkinoja-Salonen, M.S. Ionophoretic properties and mitochondrial effects of cereulide: The emetic toxin of B. cereus. Eur. J. Biochem. 1999, 263, 112–117. [Google Scholar] [CrossRef] [Green Version]
- Hoornstra, D.; Andersson, M.A.; Mikkola, R.; Salkinoja-Salonen, M.S. A new method for in vitro detection of microbially produced mitochondrial toxins. Toxicol. in Vitro 2003, 17, 745–751. [Google Scholar] [CrossRef]
- Bauer, T.; Sipos, W.; Stark, T.D.; Käser, T.; Knecht, C.; Brunthaler, R.; Saalmüller, A.; Hofmann, T.; Ehling-Schulz, M. First Insights into within Host Translocation of the Bacillus cereus Toxin Cereulide Using a Porcine Model. Front. Microbiol. 2018, 9, 2652. [Google Scholar] [CrossRef] [Green Version]
- Agata, N.; Ohta, M.; Mori, M.; Isobe, M. A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol. Lett. 1995, 129, 17–20. [Google Scholar] [CrossRef] [Green Version]
- Shinagawa, K.; Ueno, Y.; Hu, D.; Ueda, S.; Sugii, S.J. Mouse lethal activity of a HEp-2 vacuolation factor, cereulide, produced by Bacillus cereus isolated from vomiting-type food poisoning. Vet. Med. Sci. 1996, 58, 1027–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paananen, A.; Mikkola, R.; Sareneva, T.; Matikainen, S.; Hess, M.; Andersson, M.; Julkunen, I.; Salkinoja-Salonen, M.S.; Timonen, T. Inhibition of human natural killer cell activity by cereulide, an emetic toxin from Bacillus cereus. Clin. Exp. Immunol. 2002, 129, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Marxen, S.; Stark, T.D.; Rütschle, A.; Lücking, G.; Frenzel, E.; Scherer, S.; Ehling-Schulz, M.; Hofmann, T. Multiparametric Quantitation of the Bacillus cereus Toxins Cereulide and Isocereulides A–G in Foods. J. Agric. Food Chem. 2015, 63, 8307–8313. [Google Scholar] [CrossRef] [PubMed]
- Chouinard, C.D.; Nagy, G.; Smith, R.D.; Baker, E.S. Chapter Five—Ion Mobility-Mass Spectrometry in Metabolomic, Lipidomic, and Proteomic Analyses. Compr. Anal. Chem. 2019, 83, 123–159. [Google Scholar] [CrossRef]
- Biesta-Peters, E.G.; Reij, M.W.; Blaauw, R.H.; in’t Veld, P.H.; Rajkovic, A.; Ehling-Schulz, M.; Abee, T. Quantification of the emetic toxin cereulide in food products by liquid chromatography-mass spectrometry using synthetic cereulide as a standard. Appl. Environ. Microbiol. 2010, 76, 7466–7472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmann, E.; Fewer, D.P.; Neilan, B.A. Cyanobacterial toxins: Biosynthetic routes and evolutionary roots. FEMS Microbiol. Rev. 2013, 37, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Frenzel, E.; Kranzler, M.; Stark, T.D.; Hofmann, T.; Ehling-Schulz, M. The Endospore-Forming Pathogen Bacillus cereus Exploits a Small Colony Variant-Based Diversification Strategy in Response to Aminoglycoside Exposure. mBio 2015, 6, e01172-15/1–e01172-15/10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, T.; Stark, T.; Hofmann, T.; Ehling-Schulz, M. Development of a stable isotope dilution analysis for the quantification of the Bacillus cereus toxin cereulide in foods. J. Agric. Food Chem. 2010, 58, 1420–1428. [Google Scholar] [CrossRef]
- Ohmori, T.; Mutaguchi, Y.; Doi, K.; Ohshima, T. Effects of alkali or acid treatment on the isomerization of amino acids. J. Biosci. Bioeng. 2012, 114, 457–459. [Google Scholar] [CrossRef]
- Hoye, T.R.; Jeffrey, C.S.; Shao, F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nature protocols 2007, 2, 2451–2458. [Google Scholar] [CrossRef]
- Lücking, G.; Dommel, M.K.; Scherer, S.; Fouet, A.; Ehling-Schulz, M. Cereulide synthesis in emetic Bacillus cereus is controlled by the transition state regulator AbrB, but not by the virulence regulator PlcR. Microbiology 2009, 155, 922–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frenzel, E.; Doll, V.; Pauthner, M.; Lücking, G.; Scherer, S.; Ehling-Schulz, M. CodY orchestrates the expression of virulence determinants in emetic Bacillus cereus by impacting key regulatory circuits. Mol. Microbiol. 2012, 85, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Stark, T.D.; Ranner, J.; Stiglbauer, B.; Weiss, P.; Stark, S.; Balemba, O.B.; Hofmann, T. Construction and Application of a Database for a Five-Dimensional Identification of Natural Compounds in Garcinia Species by Means of UPLC-ESI-TWIMS-TOF-MS: Introducing Gas Phase Polyphenol Conformer Drift Time Distribution Intensity Ratios. J. Agric. Food Chem. 2019, 67, 975–985. [Google Scholar] [CrossRef] [PubMed]





| No. a | Variant b | F No. c | UPLC-ESI-TOF-MS Data | Structural Modification i | ||||
|---|---|---|---|---|---|---|---|---|
| RT d (min) | EM/AM e (var., ppm) | EC f mass g (Da) | Observed CCS h (Å2) | Cereulide | Variant | |||
| - | Isocereulide C | I-4, II-4 | 2.52 | 1191.66128/1191.6617 (−0.9) | C57H96N6O19Na (+16) | 355.9 | d-Ala | d-Ser |
| 6 | Isocereulide L | II-4 | 2.76 | 1191.66128/1991.6624 (−0.3) | C57H96N6O19Na (+16) | 354.8 | d-O-Leu-d-Ala | d-O-Ile-d-Ser |
| 8 | Isocereulide N | II-4 | 2.76 | 1191.66128/1991.6624 (−0.3) | C57H96N6O19Na (+16) | 354.8 | d-O-Leu d-Ala | d-O-Ile d-Ser |
| - | Isocereulide D | I-6 | 2.88 | 1147.6366/1147.6381 (+1.3) | C55H92N6O18Na (–28) | 350.3 | l-Val | l-Ala |
| 2 | Isocereulide H | II-6, II-7 | 2.88 | 1161.6522/1661.6527 (+0.4) | C56H94N6O18Na (–14) | 353.5 | d-Ala | Gly |
| 7 | Isocereulide M | II-6, II-7 | 3.07 | 1161.6522/1661.6537 (+1.3) | C56H94N6O18Na (–14) | 347.8 | d-O-Leu d-Ala | d-O-Ile Gly |
| 3 | Isocereulide I | I-6 | 3.40 | 1161.6522/1161.6538 (+1.4) | C56H94N6O18Na (–14) | 354.3 | l-Val | l-2-Abu |
| 4 | Isocereulide J | I-6 | 3.57 | 1147.6366/1147.6364 (−0.2) | C55H92N6O18Na (–28) | 347.4 | l-O-Val | l-O-Ala |
| - | Isocereulide B | II-7 | 3.83 | 1161.6522/1161.6523 (+0.1) | C56H94N6O18Na (–14) | 351.2 | d-O-Leu | d-O-Val |
| - | Isocereulide E | II-7 | 3.83 | 1161.6522/1161.6523 (+0.1) | C56H94N6O18Na (–14) | 350.1 | d-O-Leu | O-Ile |
| 1 | Cereulide | I-8, II-8 | 4.25 | 1175.6679/1175.6677 (−0.2) | C57H96N6O18Na (–) | 354.6 | - | - |
| - | Isocereulide G | I-9, II-9 | 4.56 | 1175.6679/1175.6676 (−0.3) | C57H96N6O18Na (–) | 352.4 | CI | |
| 5 | Isocereulide K | I-9, II-9 | 4.59 | 1189.6835/1189.6842 (+0.6) | C58H98N6O18Na (+14) | 361.1 | l-O-Val | l-O-Leu |
| - | Isocereulide A | I-9, II-9 | 4.83 | 1189.6835/1189.6841 (+0.5) | C58H98N6O18Na (+14) | 359.6 | l-O-Val | l-O-Ile |
| - | Isocereulide F | I-9 | 5.00 | 1189.6835/1189.6843 (+0.7) | C58H98N6O18Na (+14) | 358.1 | d-O-Leu-d-Ala | l-O-Val-l-Val |
| Variant a | Number of b | Peptide Ratio c | Additional Dipeptide | ||||
|---|---|---|---|---|---|---|---|
| d-O-Leu-d-Ala | l-O-Val-l-Val | AM (m/z) d (var., ppm) | EM (m/z) e | EC f | Sequence | ||
| Cereulide (1) | 3 | 3 | 0.78 | ||||
| Isocereulide H (2) | 2 | 3 | 0.24 | 188.0921 (−1.1) | 188.0923 | C8H14NO4 | d-O-Leu-Gly |
| Isocereulide I (3) | 3 | 2 | 0.66 | 202.1080 (+0.5) | 202.1079 | C9H16NO4 | l-O-Val-l-2-Abu |
| Isocereulide J (4) | 3 | 2 | 0.89 | 188.0925 (+1.1) | 188.0923 | C8H14NO4 | l-O-Ala-l-Val |
| Isocereulide K (5) | 3 | 2 | 1.23 | 230.1397 (+2.2) | 230.1392 | C11H20NO4 | l-O-Leu-l-Val |
| Isocereulide L g (6) | 2 | 3 | 0.30 | 218.1023 (−2.3) | 218.1028 | C9H16NO5 | d-O-Ile-d-Ser |
| Isocereulide M (7) | 1 | 3 | 0.24 | 188.0920 (−1.6) 202.1078 (−0.5) | 188.0923 202.1079 | C8H14NO4 C9H16NO4 | d-O-Leu-Gly d-O-Ile-d-Ala |
| Isocereulide N g (8) | 1 | 3 | 0.30 | 218.1023 (−2.3) 202.1076 (−1.5) | 218.1028 202.1079 | C9H16NO5 C9H16NO4 | d-O-Leu-d-Ser d-O-Ile-d-Ala |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walser, V.; Kranzler, M.; Dawid, C.; Ehling-Schulz, M.; Stark, T.D.; Hofmann, T.F. Bacillus cereus Toxin Repertoire: Diversity of (Iso)cereulide(s). Molecules 2022, 27, 872. https://doi.org/10.3390/molecules27030872
Walser V, Kranzler M, Dawid C, Ehling-Schulz M, Stark TD, Hofmann TF. Bacillus cereus Toxin Repertoire: Diversity of (Iso)cereulide(s). Molecules. 2022; 27(3):872. https://doi.org/10.3390/molecules27030872
Chicago/Turabian StyleWalser, Veronika, Markus Kranzler, Corinna Dawid, Monika Ehling-Schulz, Timo D. Stark, and Thomas F. Hofmann. 2022. "Bacillus cereus Toxin Repertoire: Diversity of (Iso)cereulide(s)" Molecules 27, no. 3: 872. https://doi.org/10.3390/molecules27030872