Discovery of New Glucose Uptake Inhibitors as Potential Anticancer Agents by Non-Radioactive Cell-Based Assays
Abstract
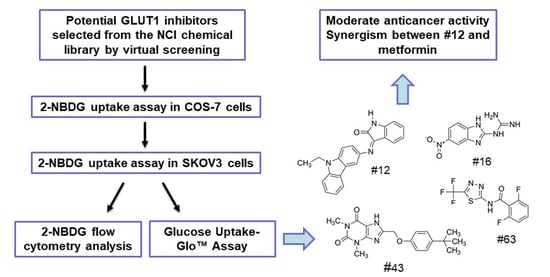
:1. Introduction
2. Results
2.1. Screening of Potential GLUT1 Inhibitors by High-Throughput 2-NBDG Uptake Assay
2.2. Validation of the Potential GLUT1 Inhibitors by Flow Cytometry Assay
2.3. Validation of the Potential GLUT1 Inhibitors by Glucose Uptake-Glo™ Assay
2.4. Comparison of Different Glucose Uptake Assay Methods in COS-7, SKOV3 and MCF-7 Cells
2.5. Effects of Potential GLUT1 Inhibitors on the Growth of SKOV3 and MCF-7 Cells
2.6. The Combination of #12 and Metformin Synergistically Inhibits SKOV3 Cell Growth
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. RT-PCR
4.3. 2-NBDG Uptake Assay
4.4. 2-NBDG Uptake Assay by Flow Cytometry
4.5. Glucose Uptake-GloTM Assay
4.6. Cell Viability Assay and Combination Index Analysis
4.7. Colony Formation Assay
4.8. Apoptosis Assay by Annexin V-FITC/PI Double Staining
4.9. Wound Healing Cell Migration Assay
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelicano, H.; Martin, D.S.; Xu, R.H.; Huang, P. Glycolysis inhibition for anticancer treatment. Oncogene 2006, 25, 4633–4646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, R.J. Glucose metabolism and cancer. Curr. Opin. Cell Biol. 2006, 18, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.B.; Figueroa, A.; Pulido, E.G.; Campelo, R.G.; Aparicio, L.A. Potential role of sugar transporters in cancer and their relationship with anticancer therapy. Int. J. Endocrinol. 2010, 2010, 205357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granchi, C.; Fortunato, S.; Minutolo, F. Anticancer agents interacting with membrane glucose transporters. MedChemComm 2016, 7, 1716–1729. [Google Scholar] [CrossRef] [Green Version]
- Reckzeh, E.S.; Waldmann, H. Development of glucose transporter (GLUT) inhibitors. Eur. J. Org. Chem. 2020, 2020, 2321–2329. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Cao, Y.; Zhang, W.; Bergmeier, S.; Qian, Y.; Akbar, H.; Colvin, R.; Ding, J.; Tong, L.; Wu, S.; et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol. Cancer Ther. 2012, 11, 1672–1682. [Google Scholar] [CrossRef] [Green Version]
- Ung, P.M.; Song, W.; Cheng, L.; Zhao, X.; Hu, H.; Chen, L.; Schlessinger, A. Inhibitor Discovery for the Human GLUT1 from Homology Modeling and Virtual Screening. ACS Chem. Biol. 2016, 11, 1908–1916. [Google Scholar] [CrossRef] [Green Version]
- Shiebeneicher, H.; Cleve, A.; Rehwinkel, H.; Neuhaus, R.; Heisler, I.; Muller, T.; Bauwer, M.; Buchmann, B. Identification and optimization of the first selective GLUT1 inhibitor BAY-876. ChemMedChem 2016, 11, 2261–2271. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Saito, M.; Matsuoka, H.; Inagaki, N. A real-time method of imaging glucose uptake in single, living mammalian cells. Nat. Protoc. 2007, 2, 753–762. [Google Scholar] [CrossRef]
- Hassanein, M.; Weidow, B.; Koehler, E.; Bakane, N.; Garbett, S.; Shyr, Y.; Quaranta, V. Development of high-throughput quantitative assays for glucose uptake in cancer cell lines. Mol. Imaging Biol. 2011, 13, 840–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reckzeh, E.S.; Karageorgis, G.; Schwalfenberg, M.; Ceballos, J.; Nowacki, J.; Stroet, M.C.M.; Binici, A.; Knauer, L.; Brand, S.; Choidas, A.; et al. Inhibition of glucose transporters and glutaminase synergistically impairs tumor cell growth. Cell Chem. Biol. 2019, 26, 1214–1228. [Google Scholar] [CrossRef] [PubMed]
- Valley, M.P.; Karassina, N.; Aoyama, N.; Carlson, C.; Cali, J.J.; Vidugiriene, J. A bioluminescent assay for measuring glucose uptake. Anal. Biochem. 2016, 505, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Takahashi, H.; Homma, T.; Saito, M.; Oh, K.B.; Nemoto, Y.; Matsuoka, H. A novel fluorescent derivative of glucose applicable to the assessment of glucose uptake activity of Escherichia coli. Biochim. Biophys. Acta 1996, 1289, 5–9. [Google Scholar] [CrossRef]
- Zou, C.; Wang, Y.; Shen, Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J. Biochem. Biophys. Methods 2005, 64, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhao, Y.; Lyu, S.; Gao, G.; Gao, Y. Identification of novel inhibitors of GLUT1 by virtual screening and cell-based assays. Investig. New Drugs 2021, 39, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Xu, C.; Sun, P.; Wu, J.; Yan, C.; Hu, M.; Yan, N. Crystal structure of the human glucose transporter GLUT1. Nature 2014, 510, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Almahmoud, S.; Wang, X.; Vennerstrom, J.L.; Zhong, H.A. Conformational Studies of glucose transporter 1 (GLUT1) as an anticancer drug target. Molecules 2019, 24, 2159. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.C.; Yang, S.F.; Huang, C.C.; Lin, C.S.; Liang, P.H.; Lin, C.J.; Hsu, L.C. Development of a novel non-radioactive cell-based method for the screening of SGLT1 and SGLT2 inhibitors using 1-NBDG. Mol. Biosyst. 2013, 9, 2010–2020. [Google Scholar] [CrossRef]
- Kao, T.Y.; Wu, H.W.; Lee, S.S.; Liang, P.H.; Guh, J.H.; Hsu, L.C. Characterization of a fluorescent glucose derivative 1-NBDG and its application in the identification of natural SGLT1/2 inhibitors. J. Food Drug Anal. 2021, 29, 521–532. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, W.; Idowu, M.O.; Oh, U.; Wang, X.Y.; Temkin, S.M.; Fang, X. Ovarian cancer relies on glucose transporter 1 to fuel glycolysis and growth: Anti-tumor activity of BAY-876. Cancers 2019, 11, 33. [Google Scholar] [CrossRef] [Green Version]
- Ben Sahra, I.; Laurent, K.; Giuliano, S.; Larbret, F.; Ponzio, G.; Gounon, P.; Marchand-Brustel, Y.L.; Giorgetti-Peraldi, S.; Cormont, M.; Bertolotto, C.; et al. Targeting cancer cell metabolism: The combination of metformin and 2-Deoxyglucose induces p53-Dependent apoptosis in prostate cancer cells. Cancer Res. 2010, 70, 2465–2475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, T.; Kang, D.; Ji, D.; Wang, X.; Zhan, W.; Fu, M.; Xin, H.B.; Wang, J.B. How does cancer cell metabolism affect tumor migration and invasion? Cell Adh. Migr. 2013, 7, 395–403. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, T.; Verdone, J.E.; Huang, J.; Kahlert, U.D.; Hernandez, J.R.; Torga, G.; Zarif, J.C.; Epstein, T.; Gatenby, R.; McCartney, A.; et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget 2015, 6, 130–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Zheng, Y.; Zhang, H.; Sun, H. Targeting cancer cell metabolism: The combination of metformin and 2-Deoxyglucose regulates apoptosis in ovarian cancer cells via p38 MAPK/JNK signaling pathway. Am. J. Transl. Res. 2016, 8, 4812–4821. [Google Scholar]
- Kapoor, K.; Finer-Moore, J.S.; Pedersen, B.P.; Caboni, L.; Waight, A.; Hillig, R.C.; Bringmann, P.; Heisler, I.; Müller, T.; Siebeneicher, H.; et al. Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides. Proc. Natl. Acad. Sci. USA 2016, 113, 4711–4716. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Heidenreich, D.; Zhou, S.; Ackloo, S.; Kramer, A.; Nakka, K.; Lima-Fernandes, E.; Deblois, G.; Duan, S.; Vellanki, R.N.; et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat. Commun. 2019, 10, 1915. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Meng, Y.Q.; Xu, X.F.; Gu, J. Blockade of GLUT1 by WZB117 resensitizes breast cancer cells to adriamycin. Anticancer Drugs 2017, 28, 880–887. [Google Scholar] [CrossRef]
- Li, Y.L.; Weng, H.C.; Hsu, J.L.; Lin, S.W.; Guh, J.H.; Hsu, L.C. The combination of MK-2206 and WZB117 exerts a synergistic cytotoxic effect against breast cancer cells. Front. Pharmacol. 2019, 10, 1311. [Google Scholar] [CrossRef] [Green Version]
- Xintaropoulou, C.; Ward, C.; Wise, A.; Marston, H.; Turnbull, A.; Langdon, S.P. A comparative analysis of inhibitors of the glycolysis pathway in breast and ovarian cancer cell line models. Oncotarget 2015, 6, 25677–25695. [Google Scholar] [CrossRef] [Green Version]
- Xintaropoulou, C.; Ward, C.; Wise, A.; Queckborner, S.; Turnbull, A.; Michie, C.O.; Williams, A.R.W.; Rye, T.; Gourley, C.; Langdon, S.P. Expression of glycolytic enzymes in ovarian cancers and evaluation of the glycolytic pathway as a strategy for ovarian cancer treatment. BMC Cancer 2018, 18, 636. [Google Scholar] [CrossRef] [PubMed]
- Sage, J.M.; Cura, A.J.; Lloyd, K.; Carruthers, A. Caffeine inhibits glucose transport by binding at the GLUT1 nucleotide-binding site. Am. J. Physiol. Cell Physiol. 2015, 308, C827–C834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, H.C.; Sung, C.J.; Hsu, J.L.; Leu, W.J.; Guh, J.H.; Kung, F.L.; Hsu, L.C. The combination of a novel GLUT1 inhibitor and cisplatin synergistically inhibits breast cancer cell growth by enhancing the DNA damaging effect and modulating the Akt/mTOR and MAPK signaling pathways. Front. Pharmacol. 2022, 13, 879748. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Nakata, M.; Horimoto, N.; Saito, M.; Matsuoka, H.; Inagaki, N. Measurement of glucose uptake and intracellular calcium concentration in single, living pancreatic β-cells. J. Biol. Chem. 2000, 275, 22278–22283. [Google Scholar] [CrossRef] [Green Version]
- Blodgett, A.B.; Kothinti, R.K.; Kamyshko, I.; Petering, D.H.; Kumar, S.; Tabatabai, N.M. A fluorescence method for measurement of glucose transport in kidney cells. Diabetes Technol. Ther. 2011, 13, 743–751. [Google Scholar] [CrossRef] [Green Version]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]






| % Uptake in COS-7 a | 2-NBDG | 2DG |
| Phl | 53.83 ± 0.87 | 8.72 ± 3.22 |
| #12 | 77.46 ± 3.82 | 86.04 ± 1.06 |
| #16 | 74.36 ± 2.99 | 71.27 ± 1.40 |
| #43 | 59.68 ± 2.28 | 6.38 ± 2.36 |
| #69 | 76.96 ± 5.91 | 15.24 ± 3.46 |
| % Uptake in SKOV3 a | 2-NBDG | 2DG |
| Phl | 55.58 ± 2.87 | 3.81 ± 1.35 |
| #12 | 56.46 ± 8.05 | 88.06 ± 10.40 |
| #16 | 62.07 ± 2.17 | 77.11 ± 7.40 |
| #43 | 67.66 ± 3.93 | 50.03 ± 3.34 |
| #69 | 69.87 ± 0.72 | 67.96 ± 7.82 |
| % Uptake in MCF-7 b | 2-NBDG flow | 2DG |
| Phl | 36.87 ± 1.31 | 10.32 ± 2.76 |
| #12 | 75.96 ± 1.75 | 101.07 ± 4.16 |
| #16 | 77.28 ± 6.21 | 98.07 ± 8.12 |
| #43 | 35.84 ± 1.11 | 29.24 ± 2.52 |
| #69 | 61.81 ± 6.14 | 104.18 ± 4.41 |
| WZB | 27.35 ± 0.98 | 2.43 ± 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, H.-C.; Li, L.-C.; Guh, J.-H.; Kung, F.-L.; Hsu, L.-C. Discovery of New Glucose Uptake Inhibitors as Potential Anticancer Agents by Non-Radioactive Cell-Based Assays. Molecules 2022, 27, 8106. https://doi.org/10.3390/molecules27228106
Hung H-C, Li L-C, Guh J-H, Kung F-L, Hsu L-C. Discovery of New Glucose Uptake Inhibitors as Potential Anticancer Agents by Non-Radioactive Cell-Based Assays. Molecules. 2022; 27(22):8106. https://doi.org/10.3390/molecules27228106
Chicago/Turabian StyleHung, Hsueh-Chih, Li-Cheng Li, Jih-Hwa Guh, Fan-Lu Kung, and Lih-Ching Hsu. 2022. "Discovery of New Glucose Uptake Inhibitors as Potential Anticancer Agents by Non-Radioactive Cell-Based Assays" Molecules 27, no. 22: 8106. https://doi.org/10.3390/molecules27228106