Development of Semisynthetic Apoptosis-Inducing Agents Based on Natural Phenolic Acids Scaffold: Design, Synthesis and In-Vitro Biological Evaluation
Abstract
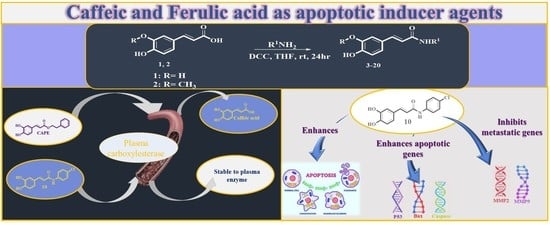
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Activity
2.2.1. Cell Viability Determination by MTT Assay
2.2.2. Cell Cycle Progression
Flow Cytometry Analysis Showing Induction of Apoptosis
Evaluation of Apoptosis by Annexin V-FITC Staining
2.2.3. Gene Expression
Caffeic Acid and Ferulic Acid Derivatives Enhanced Pro-Apoptotic Genes
Caffeic Acid and Ferulic Acid Derivatives Inhibit Metastasis Enhancing Genes
2.3. Structure–Activity Relationship Correlation
3. Materials and Methods
3.1. General
3.2. Plant Material
Extraction and Isolation of Caffeic and Ferulic Acids
3.3. Chemistry:
3.3.1. General method of Preparation of Caffeic or Ferulic Acid Amide Derivatives (3–20)
(E)-3-(3,4-Dihydroxyphenyl)-1-(4-phenylpiperazin-1-yl)prop-2-en-1-one (3)
(E)-3-(3,4-Dihydroxyphenyl)-N-(4-hydroxyphenyl)acrylamide (4)
(E)-3-(3,4-Dihydroxyphenyl)-N-(p-tolyl)acrylamide (5)
(E)-N-Cyclohexyl-3-(3,4-dihydroxyphenyl)acrylamide (6)
(E)-N-(4-Bromophenyl)-3-(3,4-dihydroxyphenyl)acrylamide (7)
(E)-N-(4-Methoxyphenyl)-3-(3,4-dihydroxyphenyl)acrylamide (8)
(E)-N-Benzyl-3-(3,4-dihydroxyphenyl)acrylamide (9)
(E)-N-(4-Chlorophenyl)-3-(3,4-dihydroxyphenyl)acrylamide (10)
(E)-N-(2-Aminophenyl)-3-(3,4-dihydroxyphenyl)acrylamide (11)
(E)-3-(3-Hydroxy-4-methoxyphenyl)-1-(4-phenylpiperazin-1-yl)prop-2-en-1-one (12)
(E)-3-(3-Hydroxy-4-methoxyphenyl)-N-(4-methoxyphenyl)acrylamide (13)
(E)-3-(3-Hydroxy-4-methoxyphenyl)-N-(p-tolyl)acrylamide (14)
(E)-N-Cyclohexyl-3-(4-hydroxy-3-methoxyphenyl)acrylamide (15)
(E)-3-(4-Hydroxy-3-methoxyphenyl)-N-(4-methoxyphenyl)acrylamide (16)
(E)-3-(3-Hydroxy-4-methoxyphenyl)-N-(m-tolyl)acrylamide (17)
(E)-N-Benzyl-3-(3-hydroxy-4-methoxyphenyl) acrylamide (18)
(E)-N-(4-Chlorophenyl)-3-(4-hydroxy-3-methoxyphenyl)acrylamide (19)
(E)-N-(2-Aminophenyl)-3-(3-hydroxy-4-methoxyphenyl)acrylamide (20)
3.4. Biological Activity
3.4.1. Cell Culture
3.4.2. Cell Viability Determination by MTT Assay
3.4.3. Annexin V/Propidium Iodide (PI) Apoptosis Assay by Flow Cytometry
3.4.4. Quantitative Polymerase Chain Reaction (qPCR) and RNA Extraction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Mattiuzzi, C.; Lippi, G. Current Cancer Epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, A.; Laakso, I.; Seppänen-Laakso, T.; Huhtikangas, A.; Riekkola, M.L. A simplified procedure for indole alkaloid extraction from Catharanthus roseus combined with a semi-synthetic production process for vinblastine. Molecules 2007, 12, 1307–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichota, A.; Gwozdzinski, K. Anticancer Activity of Natural Compounds from Plant and Marine Environment. Int. J. Mol. Sci. 2018, 19, 3533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canel, C.; Moraes, R.M.; Dayan, F.E.; Ferreira, D. Podophyllotoxin. Phytochemistry 2000, 54, 115–120. [Google Scholar] [CrossRef]
- Gomes, C.A.; da Cruz, T.G.; Andrade, J.L.; Milhazes, N.; Borges, F.; Marques, M.P.M. Anticancer activity of phenolic acids of natural or synthetic origin: A structure−activity study. J. Med. Chem. 2003, 46, 5395–5401. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Mumper, R.J. Plant phenolics: Extraction, analysis and their antioxidant and anticancer properties. Molecules 2010, 15, 7313–7352. [Google Scholar] [CrossRef]
- Panwar, R.; Sharma, A.K.; Kaloti, M.; Dutt, D.; Pruthi, V. Characterization and anticancer potential of ferulic acid-loaded chitosan nanoparticles against ME-180 human cervical cancer cell lines. Appl. Nanosci. 2016, 6, 803–813. [Google Scholar] [CrossRef] [Green Version]
- Elansary, H.O.; Szopa, A.; Kubica, P.; Al-Mana, F.A.; Mahmoud, E.A.; El-Abedin, T.K.A.Z.; Mattar, M.A.; Ekiert, H. Phenolic compounds of Catalpa speciosa, Taxus cuspidata, and Magnolia acuminata have antioxidant and anticancer activity. Molecules 2019, 24, 412. [Google Scholar] [CrossRef] [Green Version]
- Genaro-Mattos, T.C.; Maurício, Â.Q.; Rettori, D.; Alonso, A.; Hermes-Lima, M. Antioxidant activity of caffeic acid against iron-induced free radical generation—A chemical approach. PLoS ONE 2015, 10, e0129963. [Google Scholar]
- Kilani-Jaziri, S.; Mokdad-Bzeouich, I.; Krifa, M.; Nasr, N.; Ghedira, K.; Chekir-Ghedira, L. Immunomodulatory and cellular anti-oxidant activities of caffeic, ferulic, and p-coumaric phenolic acids: A structure–activity relationship study. Drug Chem. Toxicol. 2017, 40, 416–424. [Google Scholar] [CrossRef]
- Kadar, N.N.M.A.; Ahmad, F.; Teoh, S.L.; Yahaya, M.F. Caffeic Acid on Metabolic Syndrome: A Review. Molecules 2021, 26, 5490. [Google Scholar] [CrossRef] [PubMed]
- Maity, S.; Kinra, M.; Nampoothiri, M.; Arora, D.; Pai, K.S.R.; Mudgal, J. Caffeic acid, a dietary polyphenol, as a promising candidate for combination therapy. Chem. Pap. 2022, 76, 1271–1283. [Google Scholar] [CrossRef]
- Lin, Y.; Yan, Y. Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb. Cell Factories 2012, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Lin, Y.; Yan, Y. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 2013, 110, 3188–3196. [Google Scholar] [CrossRef] [PubMed]
- Tošović, J. Spectroscopic features of caffeic acid: Theoretical study. Kragujev. J. Sci. 2017, 39, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Agunloye, O.M.; Oboh, G.; Ademiluyi, A.O.; Ademosun, A.O.; Akindahunsi, A.A.; Oyagbemi, A.A.; Omobowale, T.O.; Ajibade, T.O.; Adedapo, A.A. Cardio-protective and antioxidant properties of caffeic acid and chlorogenic acid: Mechanistic role of angiotensin converting enzyme, cholinesterase and arginase activities in cyclosporine induced hypertensive rats. Biomed. Pharmacother. 2019, 109, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.P.; Hansch, C. An approach towards the quantitative structure–activity relationships of caffeic acid and its derivatives. ChemBioChem 2004, 5, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.; Araujo, R.G.; Prather, K.; Kluskens, L.; Rodrigues, L. Heterologous production of caffeic acid from tyrosine in Escherichia coli. Enzym. Microb. Technol. 2015, 71, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Sznitowska, M.; Janicki, S.; Dabrowska, E.A.; Gajewska, M. Physicochemical screening of antimicrobial agents as potential preservatives for submicron emulsions. Eur. J. Pharm. Sci. 2002, 15, 489–495. [Google Scholar] [CrossRef]
- Xie, J.; Yang, F.; Zhang, M.; Lam, C.; Qiao, Y.; Xiao, J.; Zhang, D.; Ge, Y.; Fu, L.; Xie, D. Antiproliferative activity and SARs of caffeic acid esters with mono-substituted phenylethanols moiety. Bioorganic Med. Chem. Lett. 2017, 27, 131–134. [Google Scholar] [CrossRef]
- Bispo, V.S.; Dantas, L.S.; Filho, A.D.B.C.; Pinto, I.F.; Da Silva, R.P.; Otsuka, F.A.M.; Santos, R.B.; Santos, A.C.; Trindade, D.J.; Matos, H.R. Reduction of the DNA damages, hepatoprotective effect and antioxidant potential of the coconut water, ascorbic and caffeic acids in oxidative stress mediated by ethanol. An. Acad. Bras. Cienc. 2017, 89, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Kang, N.J.; Kim, J.H.; Lee, K.M.; Lee, D.E.; Hur, H.J.; Lee, H.J. Caffeic acid phenethyl ester inhibits invasion and expression of matrix metalloproteinase in SK-Hep1 human hepatocellular carcinoma cells by targeting nuclear factor kappa B. Genes Nutr. 2008, 2, 319–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, C.; Lee, C.S.; Lee, J.-K.; Kim, T.-J.; Lee, K.-H.; Yang, Y.M.; Kim, Y.-N.; Ye, S.-K.; Chung, M.-H. CADPE suppresses cyclin D1 expression in hepatocellular carcinoma by blocking IL-6-induced STAT3 activation. Anticancer Res. 2010, 30, 481–488. [Google Scholar]
- Gu, W.; Yang, Y.; Zhang, C.; Zhang, Y.; Chen, L.; Shen, J.; Li, G.; Li, Z.; Li, L.; Li, Y.; et al. Caffeic acid attenuates the angiogenic function of hepatocellular carcinoma cells via reduction in JNK-1-mediated HIF-1α stabilization in hypoxia. RSC Adv. 2016, 6, 82774–82782. [Google Scholar] [CrossRef]
- Phadke, A.V.; Tayade, A.A.; Khambete, M.P. Therapeutic potential of ferulic acid and its derivatives in Alzheimer’s disease—A systematic review. Chem. Biol. Drug Des. 2021, 98, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Tee-Ngam, P.; Nunant, N.; Rattanarat, P.; Siangproh, W.; Chailapakul, O. Simple and rapid determination of ferulic acid levels in food and cosmetic samples using paper-based platforms. Sensors 2013, 13, 13039–13053. [Google Scholar] [CrossRef] [Green Version]
- Balasubashini, M.S.; Rukkumani, R.; Viswanathan, P.; Menon, V.P. Ferulic acid alleviates lipid peroxidation in diabetic rats. Phytother. Res. 2004, 18, 310–314. [Google Scholar] [CrossRef]
- Koh, P.-O. Ferulic acid prevents the cerebral ischemic injury-induced decreases of astrocytic phosphoprotein PEA-15 and its two phosphorylated forms. Neurosci. Lett. 2012, 511, 101–105. [Google Scholar] [CrossRef]
- Koh, P.-O. Ferulic acid prevents the cerebral ischemic injury-induced decrease of Akt and Bad phosphorylation. Neurosci. Lett. 2012, 507, 156–160. [Google Scholar] [CrossRef]
- Koh, P.-O. Ferulic acid attenuates the injury-induced decrease of protein phosphatase 2A subunit B in ischemic brain injury. PLoS ONE 2013, 8, e54217. [Google Scholar]
- Cheng, C.Y.; Su, S.Y.; Tang, N.Y.; Ho, T.Y.; Lo, W.Y.; Hsieh, C.L. Ferulic acid inhibits nitric oxide-induced apoptosis by enhancing GABA B1 receptor expression in transient focal cerebral ischemia in rats. Acta Pharmacol. Sin. 2010, 31, 889–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.-Y.; Lee, S.-M. Ferulic acid attenuates ischemia/reperfusion-induced hepatocyte apoptosis via inhibition of JNK activation. Eur. J. Pharm. Sci. 2012, 45, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Baskaran, N.; Manoharan, S.; Balakrishnan, S.; Pugalendhi, P. Chemopreventive potential of ferulic acid in 7, 12-dimethylbenz [a] anthracene-induced mammary carcinogenesis in Sprague–Dawley rats. Eur. J. Pharmacol. 2010, 637, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Manoharan, S.; Rajasekaran, D.; Prabhakar, M.; Karthikeyan, S.; Manimaran, A. Anti-cell proliferative efficacy of ferulic acid against 7, 12-dimethylbenz (a) anthracene induced hamster buccal pouch carcinogenesis. Asian Pac. J. Cancer Prev. 2012, 13, 5207–5211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Gong, X.; Jiang, R.; Li, H.; Du, W.; Kuang, G. Ferulic acid inhibits proliferation and promotes apoptosis via blockage of PI3K/Akt pathway in osteosarcoma cell. Am. J. Transl. Res. 2016, 8, 968. [Google Scholar]
- Kiokias, S.; Oreopoulou, V. A Review of the Health Protective Effects of Phenolic Acids against a Range of Severe Pathologic Conditions (Including Coronavirus-Based Infections). Molecules 2021, 26, 5405. [Google Scholar] [CrossRef]
- Hiorcea-Paquim, A.; Enache, T.A.; De Souza Gil, E.; Oliveira-Brett, A.M. Natural phenolic antioxidants electrochemistry: Towards a new food science methodology. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1680–1726. [Google Scholar] [CrossRef]
- Zeb, A. Concept, mechanism, and applications of phenolic antioxidants in foods. J. Food Biochem. 2020, 44, e13394. [Google Scholar] [CrossRef]
- Sato, Y.; Itagaki, S.; Kurokawa, T.; Ogura, J.; Kobayashi, M.; Hirano, T.; Sugawara, M.; Iseki, K. In vitro and in vivo antioxidant properties of chlorogenic acid and caffeic acid. Int. J. Pharm. 2011, 403, 136–138. [Google Scholar] [CrossRef]
- Zduńska, K.; Dana, A.; Kolodziejczak, A.; Rotsztejn, H. Antioxidant properties of ferulic acid and its possible application. Ski. Pharmacol. Physiol. 2018, 31, 332–336. [Google Scholar] [CrossRef]
- Kiani, R.; Arzani, A.; Maibody, S.M. Polyphenols, flavonoids, and antioxidant activity involved in salt tolerance in wheat, Aegilops cylindrica and their amphidiploids. Front. Plant Sci. 2021, 12, 493. [Google Scholar] [CrossRef] [PubMed]
- Magnani, C.; Isaac, V.L.B.; Correa, M.A.; Salgado, H.R.N. Caffeic acid: A review of its potential use in medications and cosmetics. Anal. Methods 2014, 6, 3203–3210. [Google Scholar] [CrossRef]
- Chan, W.-S.; Wen, P.-C.; Chiang, H.-C. Structure-activity relationship of caffeic acid analogues on xanthine oxidase inhibition. Anticancer Res. 1995, 15, 703–707. [Google Scholar] [PubMed]
- Son, S.; Lewis, B.A. Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: Structure—Activity relationship. J. Agric. Food Chem. 2002, 50, 468–472. [Google Scholar] [CrossRef]
- Naumowicz, M.; Kusaczuk, M.; Zając, M.; Jabłońska-Trypuć, A.; Mikłosz, A.; Gál, M.; Worobiczuk, M.; Kotyńska, J. The influence of the pH on the incorporation of caffeic acid into biomimetic membranes and cancer cells. Sci. Rep. 2022, 12, 1–15. [Google Scholar] [CrossRef]
- Chang, W.-C.; Hsieh, C.-H.; Hsiao, M.-W.; Lin, W.-C.; Hung, Y.-C.; Ye, J.-C. Caffeic acid induces apoptosis in human cervical cancer cells through the mitochondrial pathway. Taiwan J. Obstet. Gynecol. 2010, 49, 419–424. [Google Scholar] [CrossRef] [Green Version]
- Hung, M.-W.; Shiao, M.-S.; Tsai, L.-C.; Chang, G.-G.; Chang, T.-C. Apoptotic effect of caffeic acid phenethyl ester and its ester and amide analogues in human cervical cancer ME180 cells. Anticancer Res. 2003, 23, 4773–4780. [Google Scholar]
- Min, J.; Shen, H.; Xi, W.; Wang, Q.; Yin, L.; Zhang, Y.; Yu, Y.; Yang, Q.; Wang, Z.-N. Synergistic anticancer activity of combined use of caffeic acid with paclitaxel enhances apoptosis of non-small-cell lung cancer H1299 cells in vivo and in vitro. Cell. Physiol. Biochem. 2018, 48, 1433–1442. [Google Scholar] [CrossRef]
- Espíndola, K.M.M.; Ferreira, R.G.; Narvaez, L.E.M.; Rosario, A.C.R.S.; Da Silva, A.H.M.; Silva, A.G.B.; Vieira, A.P.O.; Monteiro, M.C. Chemical and pharmacological aspects of caffeic acid and its activity in hepatocarcinoma. Front. Oncol. 2019, 9, 541. [Google Scholar] [CrossRef] [Green Version]
- Nakatani, N.; Kayano, S.I.; Kikuzaki, H.; Sumino, K.; Katagiri, K.; Mitani, T. Identification, quantitative determination, and antioxidative activities of chlorogenic acid isomers in prune (Prunus domestica L.). J. Agric. Food Chem. 2000, 48, 5512–5516. [Google Scholar] [CrossRef]
- Bunzel, M.; Ralph, J.; Funk, C.; Steinhart, H. Structural elucidation of new ferulic acid-containing phenolic dimers and trimers isolated from maize bran. Tetrahedron Lett. 2005, 46, 5845–5850. [Google Scholar] [CrossRef]
- Mysliwiec, A.G.; Ornstein, D.L. Matrix metalloproteinases in colorectal cancer. Clin. Colorectal Cancer 2002, 1, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Dodurga, Y.; Eroğlu, C.; Seçme, M.; Elmas, L.; Avcı, Ç.B.; Şatıroğlu-Tufan, N.L. Anti-proliferative and anti-invasive effects of ferulic acid in TT medullary thyroid cancer cells interacting with URG4/URGCP. Tumor Biol. 2016, 37, 1933–1940. [Google Scholar] [CrossRef]
- Klassen, L.; Chequin, A.; Manica, G.; Biembengut, I.V.; Toledo, M.B.; Baura, V.A.; Pedrosa, F.D.O.; Ramos, E.; Costa, F.F.; de Souza, E.M.; et al. MMP9 gene expression regulation by intragenic epigenetic modifications in breast cancer. Gene 2018, 642, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zheng, Q.; Xing, X.; Dong, Y.; Wang, Y.; You, Y.; Chen, R.; Hu, C.; Chen, J.; Gao, D.; et al. Matrix stiffness-upregulated LOXL2 promotes fibronectin production, MMP9 and CXCL12 expression and BMDCs recruitment to assist pre-metastatic niche formation. J. Exp. Clin. Cancer Res. CR 2018, 37, 99. [Google Scholar] [CrossRef]
- Aborehab, N.M.; Elnagar, M.R.; Waly, N.E. Gallic acid potentiates the apoptotic effect of paclitaxel and carboplatin via overexpression of Bax and P53 on the MCF-7 human breast cancer cell line. J. Biochem. Mol. Toxicol. 2021, 35, e22638. [Google Scholar] [CrossRef] [PubMed]










| Cpd. No. | R | R1 | Cpd. No. | R | R1 |
|---|---|---|---|---|---|
| 3 | H |  | 12 | CH3 |  |
| 4 | H |  | 13 | CH3 |  |
| 5 | H |  | 14 | CH3 |  |
| 6 | H |  | 15 | CH3 |  |
| 7 | H |  | 16 | CH3 |  |
| 8 | H |  | 17 | CH3 |  |
| 9 | H |  | 18 | CH3 |  |
| 10 | H |  | 19 | CH3 |  |
| 11 | H |  | 20 | CH3 |  |
| Cpd. No. | IC50 (μM) ± SD | SI * | ||||
|---|---|---|---|---|---|---|
| MCF-7 | HepG2 | A549 | Caco-2 | PANC-1 | Vero | |
| Caffeic acid (1) | 0.97 ± 6.27 | 1.03 ± 12.63 | 1.63 ± 15.56 | 0.41 ± 3.3 | 1.29 ± 12.16 | 0.90 |
| Ferulic acid (2) | 1.12 ± 10.15 | 0.81 ± 2.17 | 1.74 ± 10.8 | 0.41 ± 1.92 | 1.08 ± 2.27 | 1.08 |
| Doxorubicin | 0.17 ± 2.00 | 0.18 ± 3.4 | 0.14 ± 1.3 | 0.17 ± 1.1 | 0.16 ± 1.86 | 0.04 |
| Cpd. No. | IC50 (μM) | SI Vero | Cpd. No. | IC50 (μM) | SI * Vero |
|---|---|---|---|---|---|
| 3 | 0.08 ± 0.62 | 0.087 | 12 | 0.04 ± 0.34 | 0.048 |
| 4 | 0.07 ± 0.36 | 0.094 | 13 | 0.08 ± 1.2 | 0.089 |
| 5 | 0.06 ± 0.35 | 0.34 | 14 | 0.05 ± 0.59 | 0.18 |
| 6 | 0.05 ± 0.44 | 0.083 | 15 | 0.02 ± 0.17 | 0.13 |
| 7 | 0.02 ± 0.11 | 0.99 | 16 | 0.08 ± 0.69 | 0.13 |
| 8 | 0.08 ± 0.32 | 0.14 | 17 | 0.02 ± 0.21 | 0.12 |
| 9 | 0.03 ± 0.29 | 0.10 | 18 | 0.05 ± 0.86 | 0.15 |
| 10 | 0.009 ± 0.09 | 0.13 | 19 | 0.02 ± 0.65 | 0.077 |
| 11 | 0.01 ± 0.11 | 0.13 | 20 | 0.01 ± 0.63 | 0.082 |
| Sample Data | Results | |||
|---|---|---|---|---|
| Cpd. No. | %G0–G1 | %S | %G2/M | %Pre-G1 |
| Control (Caco-2) | 53.61 ± 3.16 | 37.95 ± 2.25 | 8.44 ± 0.63 | 1.78 ± 0.08 |
| 1 | 46.29 ± 3.1 | 33.22 ± 1.99 | 20.49 ± 1.92 | 11.31 ± 0.81 |
| 2 | 39.77 ± 2.29 | 34.08 ± 2.63 | 26.15 ± 3.26 | 13.22 ± 0.63 |
| 5 | 41.52 ± 2.61 | 31.62 ± 1.79 | 26.86 ± 2.75 | 7.56 ± 0.39 |
| 7 | 28.68 ± 1.64 | 26.41 ± 2.15 | 44.91 ± 3.42 | 18.26 ± 1.42 |
| 9 | 38.51 ± 1.95 | 31.44 ± 2.87 | 30.05 ± 2.59 | 9.62 ± 0.37 |
| 10 | 29.75 ± 1.73 | 28.27 ± 1.99 | 41.98 ± 2.18 | 23.42 ± 2.12 |
| 11 | 41.09 ± 2.76 | 33.36 ± 2.73 | 25.55 ± 2.47 | 14.04 ± 0.95 |
| 14 | 34.28 ± 1.74 | 31.15 ± 3.17 | 34.57 ± 4.16 | 16.51 ± 0.63 |
| 15 | 26.59 ± 2.67 | 22.58 ± 2.91 | 50.83 ± 3.49 | 25.17 ± 3.29 |
| 17 | 42.11 ± 3.41 | 29.43 ± 3.16 | 28.46 ± 2.64 | 12.11 ± 0.97 |
| 18 | 33.26 ± 4.29 | 29.12 ± 4.05 | 37.62 ± 2.47 | 12.92 ± 2.16 |
| 20 | 39.12 ± 2.58 | 34.04 ± 2.69 | 26.84 ± 2.34 | 8.62 ± 0.69 |
| Cpd. No. | Total | Early Apoptosis | Late Apoptosis | Necrosis |
|---|---|---|---|---|
| Control (Caco-2) | 1.78 | 0.89 | 0.25 | 0.64 |
| 1 | 11.31 | 3.41 | 5.98 | 1.92 |
| 2 | 13.22 | 5.17 | 6.48 | 1.57 |
| 5 | 7.56 | 2.59 | 3.93 | 1.04 |
| 7 | 18.26 | 4.75 | 11.37 | 2.14 |
| 9 | 9.62 | 3.72 | 4.54 | 1.36 |
| 10 | 23.42 | 7.59 | 12.69 | 3.14 |
| 11 | 14.04 | 2.47 | 9.50 | 2.07 |
| 14 | 16.51 | 3.99 | 10.26 | 2.26 |
| 15 | 25.17 | 5.79 | 16.57 | 2.81 |
| 17 | 12.11 | 4.25 | 6.25 | 1.61 |
| 18 | 12.92 | 3.88 | 7.31 | 1.73 |
| 20 | 8.62 | 2.19 | 5.19 | 1.24 |
| Gene | Primers Used |
|---|---|
| p53 | F 5′-CCCCTCCTGGCCCCTGTCATCTTC-3′ R 5′-GCAGCGCCTCACAACCTCCGTCAT-3′ |
| Bcl-2 | F 5′-CCTGTGGATGACTGAGTACC-3′ R 5′-GAGACAGCCAGGAGAAATCA-3′ |
| Caspase 3 | F 5′-TTCATTATTCAGGCCTGCCGAGG-3′ R 5′-TTCTGACAGGCCATGTCATCCTCA-3′ |
| Bax | F 5′-GTTTCATCCAGGATCGAGCAG-3′ R 5′-CATCTTCTTCCAGATGGTGA-3′ |
| MMP-2 | F 5′- CAAAAACAAGAAGACATACATCTT-3′ R 5′- CAAAAACAAGAAGACATACATCTT-3′ |
| MMP-9 | F 5′- TGGGGGGCAACTCGGC-3′ R 5′- GGAATGATCTAAGCCCAG-3′ |
| β-actin | F 5′-GTGACATCCACACCCAGAGG-3′ R 5′-ACAGGATGTCAAAACTGCCC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ezzat, S.M.; Teba, H.E.S.; Shahin, I.G.; Hafez, A.M.; Kamal, A.M.; Aborehab, N.M. Development of Semisynthetic Apoptosis-Inducing Agents Based on Natural Phenolic Acids Scaffold: Design, Synthesis and In-Vitro Biological Evaluation. Molecules 2022, 27, 6724. https://doi.org/10.3390/molecules27196724
Ezzat SM, Teba HES, Shahin IG, Hafez AM, Kamal AM, Aborehab NM. Development of Semisynthetic Apoptosis-Inducing Agents Based on Natural Phenolic Acids Scaffold: Design, Synthesis and In-Vitro Biological Evaluation. Molecules. 2022; 27(19):6724. https://doi.org/10.3390/molecules27196724
Chicago/Turabian StyleEzzat, Shahira M., Heba El Sayed Teba, Inas G. Shahin, Ahmed M. Hafez, Aliaa M. Kamal, and Nora M. Aborehab. 2022. "Development of Semisynthetic Apoptosis-Inducing Agents Based on Natural Phenolic Acids Scaffold: Design, Synthesis and In-Vitro Biological Evaluation" Molecules 27, no. 19: 6724. https://doi.org/10.3390/molecules27196724