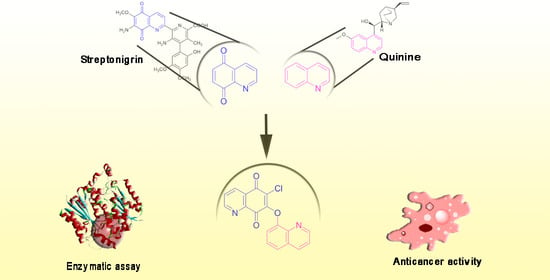
Hybrids of 1,4-Quinone with Quinoline Derivatives: Synthesis, Biological Activity, and Molecular Docking with DT-Diaphorase (NQO1)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Activity
2.2.1. Enzymatic Assay
2.2.2. Anticancer Activity
2.2.3. Apoptosis Analysis
2.3. Molecular Docking Study
3. Materials and Methods
3.1. Chemistry
- Synthesis of 2-aminoquinolin-8-ols 5–6
- Synthesis of hybrids 11–14
3.2. Biological Activity
3.2.1. Enzymatic Assay
3.2.2. Anticancer Activity
3.2.3. Apoptotic Assay
3.3. Molecular Docking
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Van de Walle, T.; Cools, L.; Mangelinckx, S.; D’hooghe, M. Recent contributions of quinolines to antimalarial and anticancer drug discovery research. Eur. J. Med. Chem. 2021, 226, 113865. [Google Scholar] [CrossRef] [PubMed]
- Akkachairin, B.; Rodphon, W.; Reamtong, O.; Mungthin, M.; Tummatorn, J.; Thongsornkleeb, C.; Ruchirawat, S. Synthesis of neocryptolepines and carbocycle-fused quinolines and evaluation of their anticancer and antiplasmodial activities. Bioorg. Chem. 2020, 98, 103732. [Google Scholar] [CrossRef] [PubMed]
- Matada, B.S.; Pattanashettar, R.; Yernale, N.G. A comprehensive review on the biological interest of quinoline and its derivatives. Bioorg. Med. Chem. 2021, 32, 115973. [Google Scholar] [CrossRef]
- Hu, Y.Q.; Gao, C.; Zhang, S.; Xu, L.; Xu, Z.; Feng, L.S.; Wu, X.; Zhao, F. Quinoline hybrids and their antiplasmodial and antimalarial activities. Eur. J. Med. Chem. 2017, 139, 22–47. [Google Scholar] [CrossRef]
- Yadav, P.; Shah, K. Quinolines, a perpetual, multipurpose scaffold in medicinal chemistry. Bioorg. Chem. 2021, 109, 104639. [Google Scholar] [CrossRef] [PubMed]
- Afzal, O.; Kumar, S.; Haider, M.R.; Ali, M.D.; Kumar, R.; Jaggi, M.; Bawa, S. A review on anticancer potential of bioactive heterocycle quinoline. Eur. J. Med. Chem. 2015, 5, 871–910. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; La Monica, G.; Bono, A.; Martorana, A. Quinoline anticancer agents active on DNA and DNA-interacting proteins: From classical to emerging therapeutic targets. Eur. J. Med. Chem. 2021, 220, 113555. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Rahi, M.; Ranjan, V.; Sharma, A. Mefloquine as a prophylaxis for malaria needs to be revisited. Int. J. Parasitol. Drugs Drug Resist. 2021, 17, 23–26. [Google Scholar] [CrossRef]
- Zhang, G.F.; Liu, X.; Zhang, S.; Pan, B.; Liu, M.L. Ciprofloxacin derivatives and their antibacterial activities. Eur. J. Med. Chem. 2018, 146, 99–612. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, H.; Yang, Y.; Chen, Z.S.; Zou, C.; Zhang, J. Chloroquine against malaria, cancers and viral diseases. Drug Discov. Today 2020, 25, 2012–2022. [Google Scholar] [CrossRef]
- Lorusso, D.; Pietragalla, A.; Mainenti, S.; Masciullo, V.; Di Vagno, G.; Scambia, G. Review role of topotecan in gynaecological cancers: Current indications and perspectives. Crit. Rev. Oncol. Hematol. 2010, 74, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Mulchin, B.J.; Newton, C.G.; Baty, J.W.; Grasso, C.H.; Martin, W.J.; Walton, M.C.; Dangerfield, E.M.; Plunkett, C.H.; Berridge, M.V.; Harper, J.L.; et al. The anti-cancer, anti-inflammatory and tuberculostatic activities of a series of 6,7-substituted-5,8-quinolinequinones. Bioorg. Med. Chem. 2010, 18, 3238–3251. [Google Scholar] [CrossRef] [PubMed]
- Kadela-Tomanek, M.; Bębenek, E.; Chrobak, E.; Boryczka, S. 5,8-Quinolinedione scaffold as a promising moiety of bioactive agents. Molecules 2019, 24, 4115. [Google Scholar] [CrossRef]
- Kadela, M.; Jastrzębska, M.; Bębenek, E.; Chrobak, E.; Latocha, M.; Kusz, J.; Książek, M.; Boryczka, S. Synthesis, structure and cytotoxic activity of mono- and dialkoxy derivatives of 5,8-quinolinedione. Molecules 2016, 21, 156. [Google Scholar] [CrossRef] [PubMed]
- Kadela-Tomanek, M.; Bębenek, E.; Chrobak, E.; Latocha, M.; Boryczka, S. Alkoxy and enediyne derivatives containing 1,4-benzoquinone subuni–s—Synthesis and antitumor activity. Molecules 2017, 22, 447. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, Q.; Sun, H.; Li, W. Novel diosgenin-1,4-quinone hybrids: Synthesis, antitumor evaluation, and mechanism studies. J. Steroid Biochem. Mol. Biol. 2021, 214, 105993. [Google Scholar] [CrossRef]
- Winski, S.; Hargreaves, R.; Butler, J.; Ross, D. A new screening system for NAD(P)H:quinone oxidoreductase (NQO1)-directed antitumor quinones: Identification of a new aziridinylbenzoquinone, RH1, as a NQO1-directed antitumor agent. Clin. Cancer Res. 1998, 4, 3083–3088. [Google Scholar]
- Siegel, D.; McGuinness, S.; Winski, S.; Ross, D. Genotype-phenotype relationships in studies of a polymorphism in NAD(P)H:quinone oxidoreductase 1. Pharmacogenet 1999, 9, 113–121. [Google Scholar] [CrossRef]
- Siegel, D.; Ross, D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radical. Biol. Med. 2000, 29, 246–253. [Google Scholar] [CrossRef]
- Ross, D.; Siegel, D. NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase), functions and pharmacogenetics. Methods Enzymol. 2004, 382, 115–144. [Google Scholar]
- Fryatt, T.; Goroski, D.; Nilson, Z.; Moody, C.; Beall, H. Novel quinolinequinone antitumor agents: Structure-metabolism studies with NAD(P)H:quinone oxidoreductase (NQO1). Bioorg. Med. Chem. Lett. 1999, 9, 2195–2198. [Google Scholar] [CrossRef]
- Ryu, C.K.; Jeong, H.J.; Lee, S.; You, H.J.; Chio, K.; Shim, J.Y.; Heo, Y.; Lee, C.O. Effects of 6-arylamine-5,8-quinolinediones and 6-chloro 7-arylo-5,8-isoquinolinediones on NAD(P)H:Quinone Oxidoreductase (NQO1) activity and their cytotoxic potential. Arch. Pharm. Res. 2001, 24, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Deng, B.; Xu, L.; Xu, X.; Wang, N.; Hu, T.; Yao, Z.; Du, J.; Yang, L.; Lei, Y.; et al. 2-Substituted 3-methylnaphtho[1,2-b]furan-4,5-diones as novel L-shaped ortho-quinone substrates for NAD(P)H:quinone oxidoreductase (NQO1). Eur. J. Med. Chem. 2014, 82, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Yasuda, M.; Mitscher, L.A.; Drake, S.D.; Kitos, P.A.; Thompson, S.C. Streptonigrin and lavendamycin partial structures. Probes for the minimum, potent pharmacophore of streptonigrin, lavendamycin, and synthetic quinoline-5,8-diones. J. Med. Chem. 1987, 30, 1918–1928. [Google Scholar] [CrossRef]
- Bolzán, A.D.; Bianchi, M.S. Genotoxicity of streptonigrin: A review. Mutat. Res. 2001, 488, 25–37. [Google Scholar] [CrossRef]
- Qiao, K.; Wan, L.; Sun, X.; Zhang, K.; Zhu, N.; Li, X.; Guo, K. Regioselective chlorination of quinoline N-oxides and isoquinoline N-oxides using PPh3/Cl3CCN. Eur. J. Org. Chem. 2016, 2016, 1606–1611. [Google Scholar] [CrossRef]
- Wu, M.Y.; Esteban, G.; Brogi, S.; Shionoya, M.; Wang, L.; Campiani, G.; Unzeta, U.; Inokuchi, T.; Butini, S.; Marco-Contelles, J. Donepezil-like multifunctional agents: Design, synthesis, molecular modeling and biological evaluation. Eur. J. Med. Chem. 2016, 121, 864–879. [Google Scholar] [CrossRef]
- Kadela-Tomanek, M.; Bębenek, E.; Chrobak, E.; Marciniec, K.; Latocha, M.; Kuśmierz, D.; Jastrzębska, M.; Boryczka, S. Betulin-1,4-quinone hybrids: Synthesis, anticancer activity and molecular docking study with NQO1 enzyme. Eur. J. Med. Chem. 2019, 177, 302–315. [Google Scholar] [CrossRef]
- Kadela-Tomanek, M.; Jastrzębska, M.; Marciniec, K.; Chrobak, E.; Bębenek, E.; Latocha, M.; Kuśmierz, D.; Boryczka, S. Design, synthesis and biological activity of 1,4-quinone moiety attached to betulin derivatives as potent DT-diaphorase substrate. Bioorg. Chem. 2021, 106, 104478. [Google Scholar] [CrossRef]
- Ling, Y.; Yang, Q.X.; Teng, Y.N.; Chen, S.; Gao, W.J.; Guo, J.; Hsu, P.L.; Liu, Y.; Morris-Natschke, S.L.; Hung, C.C.; et al. Development of novel amino-quinoline-5,8-dione derivatives as NAD(P)H:quinone oxidoreductase 1 (NQO1) inhibitors with potent antiproliferative activities. Eur. J. Med. Chem. 2018, 154, 199–209. [Google Scholar] [CrossRef]
- Modranka, J.; Drogosz-Stachowicz, J.; Pietrzak, A.; Janecka, A.; Janecki, T. Synthesis and structure-activity relationship study of novel 3-diethoxyphosphorylfuroquinoline-4,9-diones with potent antitumor efficacy. Eur. J. Med. Chem. 2021, 219, 113429. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.Q.; Ma, X.; Zhang, C.; Liu, Z.P. Design, synthesis, and biological evaluation of 4-substituted-3,4-dihydrobenzo[h]quinoline-2,5,6(1H)-triones as NQO1-directed antitumor agents. Eur. J. Med. Chem. 2020, 198, 112396. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Beyfuss, K.; Hood, D.A. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018, 23, 100–117. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative stress: Harms and benefits for human health. Oxid. Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef]
- Dlamini, Z.; Mbita, Z.; Zungu, M. Genealogy, expression, and molecular mechanisms in apoptosis. Pharmacol. Ther. 2004, 101, 1–15. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, L. The transcriptional targets of p53 in apoptosis control. Biochem. Biophys. Res. Commun. 2005, 331, 851–858. [Google Scholar] [CrossRef]
- Barr, A.R.; Cooper, S.; Heldt, F.S.; Butera, F.; Stoy, H.; Mansfeld, J.; Novak, B.; Bakal, C. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 2017, 8, 14728–14745. [Google Scholar] [CrossRef]
- Yip, K.W.; Reed, J.C. Bcl-2 family proteins and cancer. Oncogene 2008, 27, 6398–6406. [Google Scholar] [CrossRef] [PubMed]
- Woods, J.J.; Unnerstall, R.; Hasson, A.; Abou, D.S.; Radchenko, V.; Thorek, D.L.J.; WIlson, J.J. Stable chelation of the uranyl ion by acyclic hexadentate ligands: Potential applications for 230U targeted α-therapy. Inorg. Chem. 2022, 61, 3337–3350. [Google Scholar] [CrossRef]
- Batenko, N.; Popova, O.; Belyakov, S.; Valters, R. Synthesis of aminovinyl derivatives of quinoline- and isoquinoline-5,8-diones. Chem. Heterocycl. Compounds 2012, 48, 888–891. [Google Scholar] [CrossRef]
- Gupta, A.; Kumar, J.; Bhadra, S. Chelation-assisted de-aryloxylative amination of 2-aryloxy quinolines: A new synthetic route to a key fragment of a bioactive PRMT5 inhibitor. Org. Biomol. Chem. 2018, 16, 3716–3720. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bian, J.; Wang, N.; Qian, X.; Gu, J.; Mu, T.; Fan, J.; Yang, X.; Li, S.; Yang, T.; et al. Novel naphtho[2,1-d]oxazole-4,5-diones as NQO1 substrates with improved aqueous solubility: Design, synthesis, and in vivo antitumor evaluation. Bioorg. Med. Chem. 2016, 24, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Dym, O.; Tsvetkov, P.; Adler, J.; Shaul, Y. The crystal structure of NAD(P)H Quinone Oxidoreductase 1 in complex with its potent inhibitor dicoumarol. Biochemistry 2006, 45, 6372–6378. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- BIOVIA. Discovery Studio Modeling Environment; Release 2017; Dessault Systemes: San Diego, CA, USA, 2016. [Google Scholar]








| Compound | Cell Line/IC50 (µM) | ||||||
|---|---|---|---|---|---|---|---|
| Colo-829 | SK-OV-3 | MDA-MB-231 | T47D | MCF-7 | A549 | HFF-1 | |
| 11a | 18.51 ± 1.31 | 1.55 ± 0.20 | 11.49 ± 0.19 | 18.68 ± 0.71 | 19.35 ± 0.37 | 1.68 ± 0.05 | 10.21 ± 0.16 |
| 11b | 1.92 ± 0.11 | 1.69 ± 0.03 | 1.64 ± 0.15 | 2.42 ± 0.13 | 2.11 ± 0.08 | 1.78 ± 0.21 | 26.06 ± 1.02 |
| 11c | 11.65 ± 1.36 | 1.78 ± 0.31 | 14.35 ± 2.01 | 22.19 ± 0.66 | 18.33 ± 1.23 | 1.45 ± 0.13 | 10.52 ± 0.87 |
| 11d | 1.45 ± 0.24 | 1.56 ± 0.11 | 2.03 ± 0.05 | 2.10 ± 0.11 | 2.11 ± 0.09 | 1.71 ± 0.03 | 13.74 ± 1.27 |
| 11e | 15.01 ± 0.32 | 1.72 ± 0.09 | 1.97 ± 0.13 | 20.79 ± 1.03 | 2.03 ± 0.03 | 12.12 ± 1.06 | 11.45 ± 0.69 |
| 11f | 14.04 ± 1.16 | 10.89 ± 0.48 | 13.75 ± 1.09 | 18.62 ± 0.62 | 16.21 ± 0.12 | 12.63 ± 0.82 | 25.64 ± 0.19 |
| 12a | 1.52 ± 0.35 | 0.05 ± 0.02 | 1.97 ± 0.15 | 1.59 ± 0.15 | 2.09 ± 0.02 | 0.85 ± 0.04 | 17.34 ± 0.89 |
| 12b | 1.69 ± 0.08 | 1.49 ± 0.21 | 1.57 ± 0.19 | 1.90 ± 0.23 | 1.78 ± 0.03 | 1.43 ± 0.06 | 10.15 ± 1.33 |
| 12c | 14.55 ± 0.51 | 2.16 ± 0.19 | 21.12 ± 0.11 | 23.26 ± 1.12 | 19.11 ± 0.95 | 3.14 ± 0.43 | 17.89 ± 1.37 |
| 12d | 13.12 ± 0.49 | 15.38 ± 0.48 | 15.55 ± 0.79 | 16.17 ± 0.98 | 17.41 ± 0.82 | 1.46 ± 0.06 | 10.28 ± 1.65 |
| 12e | 1.53 ± 0.08 | 1.05 ± 0.06 | 1.50 ± 0.15 | 17.35 ± 1.30 | 1.89 ± 0.23 | 15.12 ± 0.39 | 17.56 ± 1.05 |
| 12f | 13.35 ± 0.21 | 13.49 ± 0.21 | 10.22 ± 0.28 | 17.34 ± 0.26 | 14.11 ± 0.67 | 11.56 ± 0.72 | 19.93 ± 0.93 |
| 13a | 0.17 ± 0.02 | 0.46 ± 0.02 | 1.02 ± 0.15 | 0.09 ± 0.04 | 1.16 ± 0.15 | 0.03 ± 0.01 | 27.69 ± 0.70 |
| 13b | 0.39 ± 0.11 | 0.49 ± 0.05 | 0.11 ± 0.01 | 1.29 ± 0.03 | 0.61 ± 0.09 | 0.57 ± 0.02 | 26.04 ± 1.60 |
| 13c | 0.34 ± 0.28 | 0.32 ± 0.01 | 0.75 ± 0.09 | 0.52 ± 0.08 | 0.89 ± 0.03 | 1.32 ± 0.32 | 13.25 ± 0.85 |
| 13d | 1.96 ± 0.08 | 17.08 ± 1.76 | 1.67 ± 0.11 | Neg. | 1.75 ± 0.19 | 0.06 ± 0.01 | 11.09 ± 1.07 |
| 13e | 19.57 ± 1.32 | 15.85 ± 0.26 | 16.61 ± 0.38 | Neg. | 16.85 ± 0.50 | 14.04 ± 0.26 | 15.52 ± 0.72 |
| 13f | 2.10 ± 0.06 | 1.23 ± 0.08 | 0.13 ± 0.05 | 2.15 ± 0.06 | 1.03 ± 0.19 | 1.22 ± 0.08 | 18.39 ± 0.35 |
| 14a | 15.30 ± 2.56 | 1.70 ± 0.29 | 15.22 ± 0.85 | Neg. | 14.07 ± 0.98 | 0.65 ± 0.07 | 12.81 ± 0.85 |
| 14b | 15.04 ± 1.03 | 16.78 ± 0.59 | 19.15 ± 0.62 | 22.16 ± 1.03 | 16.31 ± 1.08 | 2.10 ± 0.29 | 11.14 ± 1.62 |
| 14c | Neg. | 0.25 ± 0.06 | 25.14 ± 0.97 | 1.62 ± 0.21 | Neg. | 0.41 ± 0.06 | 17.96 ± 1.75 |
| 14d | 17.98 ± 2.01 | 14.12 ± 0.52 | 2.05 ± 0.35 | Neg. | 11.66 ± 1.34 | 8.13 ± 1.02 | 14.32 ± 1.23 |
| 14e | 22.00 ± 0.60 | 15.92 ± 0.22 | 7.66 ± 1.12 | Neg. | 24.05 ± 0.21 | 22.14 ± 0.89 | 18.65 ± 1.36 |
| 14f | 11.35 ± 1.97 | 14.16 ± 0.76 | 2.35 ± 0.09 | Neg. | 17.45 ± 1.02 | 14.02 ± 0.86 | 20.30 ± 0.87 |
| Doxorubicin | 0.05 ± 0.01 | 0.15 ± 0.03 | 0.64 ± 0.06 | 0.04 ± 0.01 | 0.12 ± 0.05 | 0.02 ± 0.01 | 0.12 ± 0.03 |
| Ligand | ΔG (kcal/mol) | Ligand | ΔG (kcal/mol) |
|---|---|---|---|
| 11a | −10.1 | 13a | −10.4 |
| 11b | −10.4 | 13b | −10.7 |
| 11c | −10.3 | 13c | −9.9 |
| 11d | −10.0 | 13d | −10.9 |
| 11e | −10.9 | 13e | −10.6 |
| 11f | −10.1 | 13f | −10.8 |
| 12a | −10.3 | 14a | −10.3 |
| 12b | −10.6 | 14b | −10.1 |
| 12c | −10.5 | 14c | −9.7 |
| 12d | −10.6 | 14d | −9.5 |
| 12e | −10.2 | 14e | −9.4 |
| 12f | −10.3 | 14f | −9.6 |
| ST | −7.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadela-Tomanek, M.; Jastrzębska, M.; Chrobak, E.; Bębenek, E.; Latocha, M. Hybrids of 1,4-Quinone with Quinoline Derivatives: Synthesis, Biological Activity, and Molecular Docking with DT-Diaphorase (NQO1). Molecules 2022, 27, 6206. https://doi.org/10.3390/molecules27196206
Kadela-Tomanek M, Jastrzębska M, Chrobak E, Bębenek E, Latocha M. Hybrids of 1,4-Quinone with Quinoline Derivatives: Synthesis, Biological Activity, and Molecular Docking with DT-Diaphorase (NQO1). Molecules. 2022; 27(19):6206. https://doi.org/10.3390/molecules27196206
Chicago/Turabian StyleKadela-Tomanek, Monika, Maria Jastrzębska, Elwira Chrobak, Ewa Bębenek, and Małgorzata Latocha. 2022. "Hybrids of 1,4-Quinone with Quinoline Derivatives: Synthesis, Biological Activity, and Molecular Docking with DT-Diaphorase (NQO1)" Molecules 27, no. 19: 6206. https://doi.org/10.3390/molecules27196206