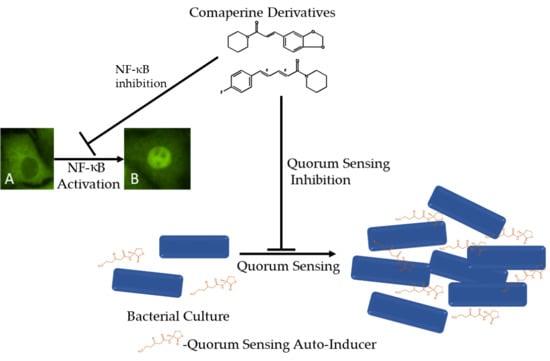
Quorum Sensing and NF-κB Inhibition of Synthetic Coumaperine Derivatives from Piper nigrum
Abstract
:1. Introduction
2. Results
2.1. Synthesis of Monoconjugated Coumaperine Derivatives
2.2. Synthesis of Di- and Triconjugated Coumaperine Derivatives
2.3. Quorum Sensing Inhibition Assay
2.4. Antibacterial Effect
2.5. NF-κB Luciferase Reporter Gene Assay
2.6. Quantitation of Activated Nuclear p65-NF-κB Fluorescence
2.7. Cell Viability Assay
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.1.1. Materials
4.1.2. Instrumentation
4.1.3. Synthesis Methods
4.2. Biological Assays
4.2.1. Quorum Sensing Inhibition Test with Bio-Reporter Chromobacterium violaceum (CV026) and Agrobacterium tumefaciens (KYC55)
4.2.2. Diffusion Discs Test
4.2.3. NF-κB-Luciferase Reporter Gene Assay
4.2.4. Quantitation of Activated Nuclear p65 Fluorescence
4.2.5. Cell Viability by XTT Assay
4.2.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- White Victoria. The Threat of Antibiotic Resistance; Drug Target Review. 2016. Available online: https://www.drugtargetreview.com/article/11525/threat-antibiotics-resistance/ (accessed on 10 April 2021).
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- WHO. Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Habboush, Y.; Guzman, N. Antibiotic Resistance. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Sitnikov, D.M.; Schineller, J.B.; Baldwin, T.O. Transcriptional Regulation of Bioluminesence Genes from Vibrio Fischeri. Mol. Microbiol. 1995, 17, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, N.A.; Barnard, A.M.L.; Slater, H.; Simpson, N.J.L.; Salmond, G.P.C. Quorum-Sensing in Gram-Negative Bacteria. FEMS Microbiol. Rev. 2001, 25, 365–404. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Kim, Y. Regulation and Controlling the Motility Properties of Pseudomonas Aeruginosa. Appl. Microbiol. Biotechnol. 2020, 104, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Moustafa, D.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The RhlR Quorum-Sensing Receptor Controls Pseudomonas Aeruginosa Pathogenesis and Biofilm Development Independently of its Canonical Homoserine Lactone Autoinducer. PLoS Pathog. 2017, 13, e1006504. [Google Scholar] [CrossRef]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A Quorum-Sensing Inhibitor Blocks Pseudomonas Aeruginosa Virulence and Biofilm Formation. Proc. Natl. Acad. Sci. USA. 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [Green Version]
- Niu, C.; Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Isolation and Characterization of an Autoinducer Synthase from Acinetobacter Baumannii. J. Bacteriol. 2008, 190, 3386–3392. [Google Scholar] [CrossRef] [Green Version]
- Gaddy, J.A.; Actis, L.A. Regulation of Acinetobacter Baumannii Biofilm Formation. Future Microbiol. 2009, 4, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Dou, Y.; Song, F.; Guo, F.; Zhou, Z.; Zhu, C.; Xiang, J.; Huan, J. Acinetobacter Baumannii Quorum-Sensing Signalling Molecule Induces the Expression of Drug-Resistance Genes. Mol. Med. Rep. 2017, 15, 4061–4068. [Google Scholar] [CrossRef] [Green Version]
- Kotas, M.E.; Medzhitov, R. Homeostasis, Inflammation, and Disease Susceptibility. Cell. 2015, 160, 816–827. [Google Scholar] [CrossRef] [Green Version]
- Khatami, M. ‘Yin and Yang’ in Inflammation: Duality in Innate Immune Cell Function and Tumorigenesis. Expert Opin. Biol. Ther. 2008, 8, 1461–1472. [Google Scholar] [CrossRef]
- Yuan, K.; Huang, C.; Fox, J.; Gaid, M.; Weaver, A.; Li, G.; Singh, B.B.; Gao, H.; Wu, M. Elevated Inflammatory Response in Caveolin-1-Deficient Mice with Pseudomonas Aeruginosa Infection is Mediated by STAT3 Protein and Nuclear Factor κB (NF-κB). J. Biol. Chem. 2011, 286, 21814–21825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golan-Goldhirsh, A.; Gopas, J. Plant Derived Inhibitors of NF-κB. Phytochem. Rev. 2014, 13, 107–121. [Google Scholar] [CrossRef]
- Pancer, Z.; Rast, J.P.; Davidson, E.H. Origins of Immunity: Transcription Factors and Homologues of Effector Genes of the Vertebrate Immune System Expressed in Sea Urchin Coelomocytes. Immunogenetics 1999, 49, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; McFadden, G. Modulation of NF-κB Signalling by Microbial Pathogens. Nature reviews. Microbiology 2011, 9, 291–306. [Google Scholar] [PubMed]
- Oliver, E.; Nandakumar, N.; Faibish, H.; Gopas, J.; Kushmaro, A. Coral-Associated Bacterial Extracts Inhibit Cellular NF-κB Pathway. Cogent Environ. Sci. 2017, 3. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The Hierarchy Quorum Sensing Network in Pseudomonas Aeruginosa. Protein Cell 2014, 6, 26–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kravchenko, V.V.; Kaufmann, G.F.; Mathison, G.C.; Scott, D.A.; Katz, A.Z.; Grauer, D.C.; Lehmann, M.; Meijler, M.M.; Janda, K.D.; Ulevitch, R.J. Modulation of Gene Expression Via Disruption of NF-κB Signaling by a Bacterial Small Molecule. Science 2008, 321, 259–263. [Google Scholar] [CrossRef]
- Peng, C.; Han, J.; Ye, X.; Zhang, X. IL-33 Treatment Attenuates the Systemic Inflammation Reaction in Acinetobacter Baumannii Pneumonia by Suppressing TLR4/NF-κB Signaling. Inflammation 2018, 41, 870–877. [Google Scholar] [CrossRef]
- Asfour, H.Z. Anti-Quorum Sensing Natural Compounds. J. Microsc. Ultrastruct. 2018, 6, 1. [Google Scholar] [CrossRef]
- Chong, Y.M.; Yin, W.F.; Ho, C.Y.; Mustafa, M.R.; Hadi, A.H.A.; Awang, K.; Narrima, P.; Koh, C.; Appleton, D.R.; Chan, K. Malabaricone C from Myristica Cinnamomea Exhibits Anti-Quorum Sensing Activity. J. Nat. Prod. 2011, 74, 2261–2264. [Google Scholar] [CrossRef]
- Packiavathy, I.A.S.V.; Priya, S.; Pandian, S.K.; Ravi, A.V. Inhibition of Biofilm Development of Uropathogens by Curcumin—An Anti-Quorum Sensing Agent from Curcuma Longa. Food Chem. 2014, 148, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, D.; Singh, V. Journal of Traditional and Complementary Medicine. J. Tradit. Complementary Med. 2016, 6, 57–61. [Google Scholar] [CrossRef] [Green Version]
- Kitano, M.; Wanibuchi, H.; Kikuzaki, H.; Nakatani, N.; Imaoka, S.; Funae, Y.; Hayashi, S.; Fukushima, S. Chemopreventive Effects of Coumaperine from Pepper on the Initiation Stage of Chemical Hepatocarcinogenesis in the Rat. Jpn. J. Cancer Res. 2000, 91, 674–680. [Google Scholar] [CrossRef]
- Muthuraman, S.; Sinha, S.; Vasavi, C.S.; Waidha, K.M.; Basu, B.; Munussami, P.; Balamurali, M.M.; Doble, M.; Saravana Kumar, R. Design, Synthesis and Identification of Novel Coumaperine Derivatives for Inhibition of Human 5-LOX: Antioxidant, Pseudoperoxidase and Docking Studies. Bioorg. Med. Chem. 2019, 27, 604–619. [Google Scholar] [CrossRef]
- Nandakumar, N.; Muthuraman, S.; Gopinath, P.; Nithya, P.; Gopas, J.; Kumar, R.S. Synthesis of Coumaperine Derivatives: Their NF-κB Inhibitory Effect, Inhibition of Cell Migration and their Cytotoxic Activity. Eur. J. Med. Chem. 2017, 125, 1076–1087. [Google Scholar] [CrossRef]
- Shingate, P.N.; Dongre, P.P.; Kannur, D.M. New Method Development for Extraction and Isolation of Piperine from Black Pepper. Int. J. Pharm. Sci. Res. 2013, 4, 3165. [Google Scholar]
- Zhu, J.; Chai, Y.; Zhong, Z.; Li, S.; Winans, S.C. Agrobacterium Bioassay Strain for Ultrasensitive Detection of N-Acylhomoserine Lactone-Type Quorum-Sensing Molecules: Detection of Autoinducers in Mesorhizobium Huakuii. Appl. Environ. Microbiol. 2003, 69, 6949–6953. [Google Scholar] [CrossRef] [Green Version]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas Aeruginosa Virulence by Quorum Sensing Inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Hossain, M.A.; Lee, S.; Park, N.; Mechesso, A.F.; Birhanu, B.T.; Kang, J.; Reza, M.A.; Suh, J.; Park, S. Impact of Phenolic Compounds in the Acyl Homoserine Lactone-Mediated Quorum Sensing Regulatory Pathways. Sci. Rep. 2017, 7, 10618. [Google Scholar] [CrossRef] [Green Version]
- Annunziato, G. Strategies to Overcome Antimicrobial Resistance (AMR) Making use of Non-Essential Target Inhibitors: A Review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef] [Green Version]
- Ozer, J.; Eisner, N.; Ostrozhenkova, E.; Bacher, A.; Eisenreich, W.; Benharroch, D.; Golan-Goldhirsh, A.; Gopas, J. Nuphar Lutea Thioalkaloids Inhibit the Nuclear Factor κappaB Pathway, Potentiate Apoptosis and are Synergistic with Cisplatin and Etoposide. Cancer Biol. Ther. 2009, 8, 1860–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joelsson, A.C.; Zhu, J. LacZ-Based Detection of Acyl-Homoserine Lactone Quorum-Sensing Signals. Curr. Protoc. Microbiol. 2006, 3, 1C.2.1–1C.2.9. [Google Scholar] [CrossRef] [PubMed]
- Küppers, R.; Engert, A.; Hansmann, M. Hodgkin Lymphoma. J. Clin. Investig. 2012, 122, 3439–3447. [Google Scholar] [CrossRef] [PubMed]











| Entry | Monoconjugated-CP | Degree of QSI of CV026 | Degree of QSI of KYC55 |
| 1. |  CP-215 | +++ | +++ |
| 2. |  CP-270 | +++ | +++ |
| 3. |  CP-296 | +++ | +++ |
| 4. |  CP-282 | +++ | ++ |
| 5. |  CP-289 | + | +++ |
| 6. |  CP-237 | ++ | ++ |
| 7. |  CP-295 | ++ | ++ |
| 8. |  CP-286 | + | ++ |
| Entry | Diconjugated-CP | Degree of QSI of CV026 | Degree of QSI of KYC55 |
| 9. |  CP-154 | +++ | +++ |
| 10. |  CP-158 | +++ | ++ |
| 11. |  CP-281 F1 | ++ | + |
| 12. |  CP-38 | ++ | + |
| 13. |  CP-9 | ++ | + |
| Entry | Triconjugated-CP | Degree of QSI of CV026 | Degree of QSI of KYC55 |
| 14. |  CP-102 | ++ | + |
| Compound Code | Bacillus subtilis (mm) | Staphylococcus aureus (mm) | Streptococcus sobrinus (mm) | Streptococcus mutans (mm) | PA01 (mm) | Acinetobacter baumannii (mm) |
|---|---|---|---|---|---|---|
| CP-147 | - | 9 | - | - | - | - |
| CP-154 | 7 | 11 | 9 | - | - | - |
| CP-9 | 8 | 9 | 9 | - | - | - |
| Compound Code | QSI | Antibacterial | NF-κB Inhibition | Cytotoxic |
|---|---|---|---|---|
| CP-9 | + | + | + | + |
| CP-154 | + | + | + | + |
| CP-158 | + | − | + | + |
| CP-215 | + | − | + | − |
| CP-286 | + | − | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadosh, Y.; Muthuraman, S.; Yaniv, K.; Baruch, Y.; Gopas, J.; Kushmaro, A.; Kumar, R.S. Quorum Sensing and NF-κB Inhibition of Synthetic Coumaperine Derivatives from Piper nigrum. Molecules 2021, 26, 2293. https://doi.org/10.3390/molecules26082293
Kadosh Y, Muthuraman S, Yaniv K, Baruch Y, Gopas J, Kushmaro A, Kumar RS. Quorum Sensing and NF-κB Inhibition of Synthetic Coumaperine Derivatives from Piper nigrum. Molecules. 2021; 26(8):2293. https://doi.org/10.3390/molecules26082293
Chicago/Turabian StyleKadosh, Yael, Subramani Muthuraman, Karin Yaniv, Yifat Baruch, Jacob Gopas, Ariel Kushmaro, and Rajendran Saravana Kumar. 2021. "Quorum Sensing and NF-κB Inhibition of Synthetic Coumaperine Derivatives from Piper nigrum" Molecules 26, no. 8: 2293. https://doi.org/10.3390/molecules26082293