Nano-SAR Modeling for Predicting the Cytotoxicity of Metal Oxide Nanoparticles to PaCa2
Abstract
:1. Introduction
2. Results and Discussion
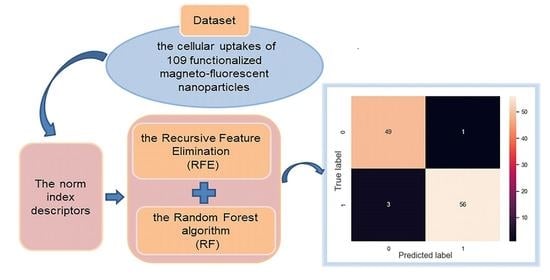
2.1. Nano-SAR Model Performance
2.2. Model Stability Validation and Results Assessment
2.3. Applicability Domain of the Proposed Model
2.4. Comparisons with Other Models in the Literature
3. Materials and Methods
3.1. Data Set
3.2. Dataset Splitting
3.3. Molecular Descriptors Calculation
3.4. Descriptor Selection and Modeling
3.5. Model Validation
3.6. Applicability Domain (AD)
4. Conclusions
- The employed norm index descriptors combining the atomic distance matrices with the property matrix could accurately and effectively characterize the structural features of MNPs and lead to a nano-SAR model with satisfactory model performance.
- The Random forest algorithm (RF) combined with the Recursive Feature Elimination (RFE) method could be successfully employed to explore and describe the internal relationships between the nanostructure and cytotoxicity of MNPs.
- Since a considerable number of MNPs were involved in the development of the model, and a rigorous model validating process and extensive model comparisons were performed, the proposed model in this study could be reasonably considered as reliable in predicting the cytotoxicity of novel MNPs or other MNPs for which experimental data are unknown.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Khojasteh, H.; Safajou, H.; Mortazavi-Derazkola, S.; Salavati-Niasari, M.; Heydaryan, K.; Yazdani, M. Economic procedure for facile and eco-friendly reduction of graphene oxide by plant extracts; a comparison and property investigation. J. Clean. Prod. 2019, 229, 1139–1147. [Google Scholar] [CrossRef]
- Mortazavi-Derazkola, S.; Salavati-Niasari, M.; Khojasteh, H.; Amiri, O.; Ghoreishi, S.M. Green synthesis of magnetic Fe3O4/SiO2/HAp nanocomposite for atenolol delivery and in vivo toxicity study. J. Clean. Prod. 2017, 168, 39–50. [Google Scholar] [CrossRef]
- Zinatloo-Ajabshir, S.; Mortazavi-Derazkola, S.; Salavati-Niasari, M. Nd2O3 nanostructures: Simple synthesis, characterization and its photocatalytic degradation of methylene blue. J. Mol. Liq. 2017, 234, 430–436. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.J.X.; Li, S.R.; Wei, H. Multifunctional nanozymes: Enzyme-like catalytic activity combined with magnetism and surface plasmon resonance. Nanoscale Horiz. 2018, 3, 367–382. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Shukla, S.; Kang, S.M.; Hwang, S.K.; Song, X.; Huh, Y.S.; Han, Y.K. Developments of cyanobacteria for nano-marine drugs: Relevance of nanoformulations in cancer therapies. Mar. Drugs 2018, 16, 179. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed, I.H.; Huang, X.H.; El-Sayed, M.A. Selective laser photo-thermal therapy of epithelial carcinoma using anti-EGFR antibody conjugated gold nanoparticles. Cancer Lett. 2006, 239, 129–135. [Google Scholar] [CrossRef]
- Dutta, T.; Kim, K.H.; Deep, A.; Szulejko, J.E.; Vellingiri, K.; Kumar, S.; Kwon, E.E.; Yun, S.T. Recovery of nanomaterials from battery and electronic wastes: A new paradigm of environmental waste management. Renew. Sust. Energ. Rev. 2018, 82, 3694–3704. [Google Scholar] [CrossRef]
- Auffan, M.; Rose, J.; Bottero, J.Y.; Lowry, G.V.; Jolivet, J.P.; Wiesner, M.R. Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. 2009, 4, 634–641. [Google Scholar] [CrossRef]
- Dong, J.Y.; Zink, J.I. Taking the temperature of the interiors of magnetically heated nanoparticles. ACS Nano 2014, 8, 5199–5207. [Google Scholar] [CrossRef]
- Jiang, S.; Gao, Q.; Chen, H.C.; Roco, M.C. The roles of sharing, transfer, and public funding in nanotechnology knowledge-diffusion networks. J. Assoc. Inf. Sci. Technol. 2015, 66, 1017–1029. [Google Scholar] [CrossRef]
- Pan, Y.; Li, T.; Cheng, J.; Telesca, D.; Zink, J.I.; Jiang, J.C. Nano-QSAR modeling for predicting the cytotoxicity of metal oxide nanoparticles using novel descriptors. RSC Adv. 2016, 6, 25766–25775. [Google Scholar] [CrossRef]
- Hansch, C.; M, P.P.; Fujita, T.; Muir, R.M. Correlation of Biological Activity of Phenoxyacetic Acids with Hammett Substituent Constants and Partition Coefficients. Nature 1962, 194, 178–180. [Google Scholar] [CrossRef]
- Winkler, D.A.; Mombelli, E.; Pietroiusti, A.; Tran, L.; Worth, A.; Fadeel, B.; McCall, M.J. Applying quantitative structure-activity relationship approaches to nanotoxicology: Current status and future potential. Toxicology 2013, 313, 15–23. [Google Scholar] [CrossRef]
- Park, S.H.; Sung, J.H.; Kim, E.J.; Chung, N. Berberine induces apoptosis via ROS generation in PANC-1 and MIA-PaCa2 pancreatic cell lines. Braz. J. Med. Biol. Res. 2015, 48, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Doi, T.; Ishikawa, T.; Okayama, T.; Oka, K.; Mizushima, K.; Yasuda, T.; Sakamoto, N.; Katada, K.; Kamada, K.; Uchiyama, K.; et al. The JAK/STAT pathway is involved in the upregulation of PD-L1 expression in pancreatic cancer cell lines. Oncol. Rep. 2017, 37, 1545–1554. [Google Scholar] [CrossRef] [Green Version]
- Hao, C.; Zhang, X.; Zhang, H.; Shang, H.; Bao, J.; Wang, H.; Li, Z. Sugiol (12-hydroxyabieta-8,11,13-trien-7-one) targets human pancreatic carcinoma cells (Mia-PaCa2) by inducing apoptosis, G2/M cell cycle arrest, ROS production and inhibition of cancer cell migration. J. Buon 2018, 23, 205–210. [Google Scholar]
- Brulle, L.; Vandamme, M.; Ries, D.; Martel, E.; Robert, E.; Lerondel, S.; Trichet, V.; Richard, S.; Pouvesle, J.-M.; Le Pape, A. Effects of a non thermal plasma treatment alone or in combination with gemcitabine in a MIA PaCa2-luc orthotopic pancreatic carcinoma model. PLoS ONE 2012, 7, e52653. [Google Scholar] [CrossRef]
- Qi, R.; Pan, Y.; Cao, J.; Jia, Z.; Jiang, J. The cytotoxicity of nanomaterials: Modeling multiple human cells uptake of functionalized magneto-fluorescent nanoparticles via nano-QSAR. Chemosphere 2020, 249. [Google Scholar] [CrossRef]
- Weissleder, R.; Kelly, K.; Sun, E.Y.; Shtatland, T.; Josephson, L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat. Biotechnol. 2005, 23, 1418–1423. [Google Scholar] [CrossRef]
- Fourches, D.; Pu, D.Q.Y.; Tassa, C.; Weissleder, R.; Shaw, S.Y.; Mumper, R.J.; Tropsha, A. Quantitative nanostructure-activity relationship modeling. ACS Nano 2010, 4, 5703–5712. [Google Scholar] [CrossRef] [Green Version]
- OECD. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] Models. 2014. Available online: http://www.oecd.org/ (accessed on 3 September 2014).
- Chau, Y.T.; Yap, C.W. Quantitative nanostructure-activity relationship modelling of nanoparticles. RSC Adv. 2012, 2, 8489–8496. [Google Scholar] [CrossRef]
- Kar, S.; Gajewicz, A.; Puzyn, T.; Roy, K. Nano-quantitative structure-activity relationship modeling using easily computable and interpretable descriptors for uptake of magnetofluorescent engineered nanoparticles in pancreatic cancer cells. Toxicol. Vitr. 2014, 28, 600–606. [Google Scholar] [CrossRef]
- Winkler, D.A.; Burden, F.R.; Yan, B.; Weissleder, R.; Tassa, C.; Shaw, S.; Epa, V.C. Modelling and predicting the biological effects of nanomaterials. SAR QSAR Environ. Res. 2014, 25, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Ojha, P.K.; Kar, S.; Roy, K.; Leszczynski, J. Toward comprehension of multiple human cells uptake of engineered nano metal oxides: Quantitative inter cell line uptake specificity (QICLUS) modeling. Nanotoxicology 2019, 13, 14–34. [Google Scholar] [CrossRef]
- Toropov, A.A.; Toropova, A.P.; Puzyn, T.; Benfenati, E.; Gini, G.; Leszczynska, D.; Leszczynski, J. QSAR as a random event: Modeling of nanoparticles uptake in PaCa2 cancer cells. Chemosphere 2013, 92, 31–37. [Google Scholar] [CrossRef]
- Singh, K.P.; Basant, N.; Gupta, S. Support vector machines in water quality management. Anal. Chim. Acta 2011, 703, 152–162. [Google Scholar] [CrossRef]
- Fjodorova, N.; Vracko, M.; Novic, M.; Roncaglioni, A.; Benfenati, E. New public QSAR model for carcinogenicity. Chem. Cent. J. 2010, 4. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Shen, J.; Yu, Y.; Li, W.; Liu, G.; Lee, P.W.; Tang, Y. In silico prediction of Tetrahymena pyriformis toxicity for diverse industrial chemicals with substructure pattern recognition and machine learning methods. Chemosphere 2011, 82, 1636–1643. [Google Scholar] [CrossRef]
- Singh, K.P.; Gupta, S. Nano-QSAR modeling for predicting biological activity of diverse nanomaterials. RSC Adv. 2014, 4, 13215–13230. [Google Scholar] [CrossRef]
- Wang, Y.L.; Yan, F.Y.; Jia, Q.Z.; Wang, Q. Assessment for multi-endpoint values of carbon nanotubes: Quantitative nanostructure-property relationship modeling with norm indexes. J. Mol. Liq. 2017, 248, 399–405. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, A. Feature selection for classifying high-dimensional numerical data. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 27 June–2 July 2004; pp. II251–II258. [Google Scholar]
- Chen, Q.; Meng, Z.P.; Liu, X.Y.; Jin, Q.G.; Su, R. Decision variants for the automatic determination of optimal feature subset in RF-RFE. Genes 2018, 9, 301. [Google Scholar] [CrossRef] [Green Version]
- Nasiri, A.; Omid, M.; Taheri-Garavand, A. An automatic sorting system for unwashed eggs using deep learning. J. Food Eng. 2020, 283, 9. [Google Scholar] [CrossRef]
- Singh, K.P.; Singh, A.K.; Gupta, S.; Rai, P. Modeling and optimization of reductive degradation of chloramphenicol in aqueous solution by zero-valent bimetallic nanoparticles. Environ. Sci. Pollut. Res. 2012, 19, 2063–2078. [Google Scholar] [CrossRef] [PubMed]
- Benigni, R.; Netzeva, T.I.; Benfenati, E.; Franke, R.; Helma, C.; Hulzebos, E.; Marchant, C.; Richard, A.M.; Woo, Y.; Yang, C. The expanding role of predictive toxicology: An update on the (Q)SAR models for mutagens and carcinogens. J. Environ. Sci. Health Part C 2007, 25, 53–97. [Google Scholar] [CrossRef]
- Singh, G.; Panda, R.K. Daily sediment yield modeling with artificial neural network using 10-fold cross validation method: A small agricultural watershed. Int. J. Earth Sci. Eng. 2011, 4, 443–450. [Google Scholar]
- Kovarich, S.; Papa, E.; Gramatica, P. QSAR classification models for the prediction of endocrine disrupting activity of brominated flame retardants. J. Hazard. Mater. 2011, 190, 106–112. [Google Scholar] [CrossRef]


| Sub-Set | n | SE | SP | ACC | MCC |
|---|---|---|---|---|---|
| Training set | 89 | 0.958 | 0.976 | 0.966 | 0.933 |
| Test set | 20 | 0.909 | 1 | 0.950 | 0.905 |
| Complete | 109 | 0.949 | 0.980 | 0.972 | 0.927 |
| Works | Method | Sub-Set | SE | SP | ACC | MCC |
|---|---|---|---|---|---|---|
| Singh et al. | DTB | Training set | 1 | 0.974 | 0.988 | 0.980 |
| Test set | 0.882 | 1 | 0.926 | 0.860 | ||
| DTF | Training set | 1 | 1 | 1 | 1 | |
| Test set | 0.875 | 0.909 | 0.889 | 0.780 | ||
| This work | RF | Training set | 0.958 | 0.976 | 0.966 | 0.933 |
| Test set | 0.909 | 1 | 0.950 | 0.905 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Pan, Y.; Yang, F.; Cao, J.; Tan, X.; Yuan, B.; Jiang, J. Nano-SAR Modeling for Predicting the Cytotoxicity of Metal Oxide Nanoparticles to PaCa2. Molecules 2021, 26, 2188. https://doi.org/10.3390/molecules26082188
Shi H, Pan Y, Yang F, Cao J, Tan X, Yuan B, Jiang J. Nano-SAR Modeling for Predicting the Cytotoxicity of Metal Oxide Nanoparticles to PaCa2. Molecules. 2021; 26(8):2188. https://doi.org/10.3390/molecules26082188
Chicago/Turabian StyleShi, Haihua, Yong Pan, Fan Yang, Jiakai Cao, Xinlong Tan, Beilei Yuan, and Juncheng Jiang. 2021. "Nano-SAR Modeling for Predicting the Cytotoxicity of Metal Oxide Nanoparticles to PaCa2" Molecules 26, no. 8: 2188. https://doi.org/10.3390/molecules26082188