Critical Protein–Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription
Abstract
:1. Introduction
2. The Serum-Response Element
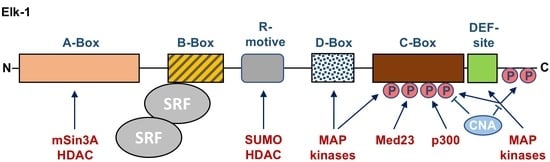
3. Modular Structure of Elk-1 and Other Ternary Complex Factors
4. Tools to Investigate Elk-1 Activity
5. Biological Functions of Elk-1
6. Essential Protein–Protein Interactions of Elk-1
6.1. Generation of a Ternary Complex with SRF: Role of DNA Binding and Protein–Protein Interactions
6.2. Phosphoacceptor Motifs of Elk-1
6.3. Docking Sites for Stimulus-Responsive MAP Kinases
6.4. Phospho-Elk-1 Interacts with the Mediator Complex
6.5. Protein–Protein Interaction of Elk-1 with the Histone Acetyltransferase p300
6.6. Calcineurin Catalyzes Dephosphorylation and Inactivation of Elk-1
6.7. Recruitment of the mSin3A–Histone Deacetylase Complex to Highly Phosphorylated Elk-1
6.8. A SUMO–Histone Deacetylase Complex Binds to Elk-1 in the Absence of Stimulation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharrocks, A.D. The ETS-domain transcription factor family. Nat. Mol. Cell Biol. 2001, 2, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.E.; Saxton, J. Ternary complex factors: Prime nuclear targets for mitogen-activated protein kinases. Int. J. Biochem. Cell Biol. 2003, 35, 1210–1226. [Google Scholar] [CrossRef]
- Yang, S.-H.; Shore, P.; Willingham, N.; Lakey, J.H.; Sharrocks, A.D. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 1999, 18, 5666–5674. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-M.; Li, L.; Papadopoulou, N.; Hodgson, G.; Evans, E.; Galbraith, M.; Dear, M.; Vougier, S.; Saxton, J.; Shaw, P.E. Mitogen-induced recruitment of ERK and MSK to SRE promoter complexes by ternary complex factor Elk-1. Nucleic Acids Res. 2008, 36, 2594–2607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tur, G.; Georgieva, E.I.; Gagete, A.; López-Rodas, G.; Rodríguez, J.L.; Franco, L. Factor binding and chromatin modification in the promoter of murine Egr1 gene upon induction. Cell. Mol. Life Sci. 2010, 67, 4065–4077. [Google Scholar] [CrossRef]
- Esnault, C.; Gualdrini, F.; Horswell, S.; Kelly, G.; Stewart, A.; East, P.; Matthews, N.; Treisman, R. ERK-induced activation of TCF family of SRF cofactors initiates a chromatin modification cascade associated with transcription. Mol. Cell 2017, 65, 1081–1095. [Google Scholar] [CrossRef] [Green Version]
- Price, M.A.; Rogers, A.E.; Treisman, R. Comparativer analysis of the ternary complex factors Elk-1, SAP-1a and SAP2 (ERP/NET). EMBO J. 1995, 14, 2589–2601. [Google Scholar] [CrossRef]
- Lesch, A.; Backes, T.M.; Langfermann, D.S.; Rössler, O.G.; Laschke, M.W.; Thiel, G. Ternary complex factor regulates pancreatic islet size and blood glucose homeostasis in transgenic mice. Pharmacol. Res. 2020, 159, 104983. [Google Scholar] [CrossRef]
- Balamotis, M.A.; Pennella, M.A.; Stevens, J.L.; Wasylyk, B.; Belmont, A.S.; Berk, A.J. Complexity in transcriptional control at the activation domain-mediator interface. Sci. Signal. 2009, 2, ra20. [Google Scholar] [CrossRef] [Green Version]
- Janknecht, R.; Ernst, W.H.; Pingoud, V.; Nordheim, A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993, 12, 5097–5104. [Google Scholar] [CrossRef]
- Marais, R.; Wynne, J.; Treisman, R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 1993, 73, 381–393. [Google Scholar] [CrossRef]
- Gille, H.; Kortenjann, M.; Thomae, O.; Moomaw, C.; Slaughter, C.; Cobb, M.H.; Shaw, P.E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995, 14, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Gille, H.; Strahl, T.; Shaw, P.E. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr. Biol. 1995, 5, 1191–1200. [Google Scholar] [CrossRef] [Green Version]
- Cavigelli, M.; Dolfi, F.; Claret, F.X.; Karin, M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 1995, 14, 5957–5964. [Google Scholar] [CrossRef] [PubMed]
- Whitmarsh, A.J.; Yang, S.-H.; Su, M.S.S.; Sharrocks, A.D.; Davis, R.J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol. Cell. Biol. 1997, 17, 2360–2371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruzalegui, F.H.; Cano, E.; Treisman, R. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene 1999, 18, 7948–7957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costello, P.S.; Nicolas, R.H.; Watanabe, Y.; Rosewell, I.; Treisman, R. Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nat. Immunol. 2004, 5, 289–298. [Google Scholar] [CrossRef]
- Cesari, F.; Brecht, S.; Vintersten, K.; Vuong, L.G.; Hofmann, M.; Klingel, K.; Schnorr, J.J.; Arsenian, S.; Schild, H.; Herdegen, T.; et al. Mice deficient for the Ets transcription factor Elk-1 show normal immune responses and mildly impaired neuronal gene activation. Mol. Cell. Biol. 2004, 24, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Ayadi, A.H.; Zheng, H.; Sobieszczuk, P.; Buchwalter, G.; Moerman, P.; Alitalo, K.; Wasylyk, B. Net-targeted mutant mice develop a vascular phenotype and up-regulate egr-1. EMBO J. 2001, 20, 5139–5152. [Google Scholar] [CrossRef] [Green Version]
- Costello, P.; Nicolas, R.; Willoughby, J.; Wasylyk, B.; Nordheim, A.; Treisman, R. Ternary complex factors SAP-1 and Elk-1, but not Net, are functionally equivalent in thymocyte development. J. Immunol. 2010, 185, 1082–1092. [Google Scholar] [CrossRef] [Green Version]
- Mayer, S.I.; Rössler, O.G.; Endo, T.; Charnay, P.; Thiel, G. Epidermal growth factor-induced proliferation of astrocytes requires Egr transcription factors. J. Cell Sci. 2009, 122, 3340–3350. [Google Scholar] [CrossRef] [Green Version]
- Vickers, E.R.; Kasza, A.; Kurnaz, I.A.; Seifert, A.; Zeef, L.A.H.; O’Donnell, A.; Hayes, A.; Sharrocks, A.D. Ternary complex factor-serum response factor complex-related gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol. Cell. Biol. 2004, 24, 10340–10351. [Google Scholar] [CrossRef] [Green Version]
- Besnard, A.; Galan-Rodriguez, B.; Vanhoutte, P.; Caboche, J. Elk-1 a transcription factor with multiple facets in the brain. Front. Neurosci. 2011, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Maurice, D.; Costello, P.; Sargent, M.; Treisman, R. ERK signaling controls innate-like CD8+ T cell differentiation via the ELK4 (SAP-1) and ELK1 transcription factors. J. Immunol. 2018, 201, 1681–1691. [Google Scholar] [CrossRef] [Green Version]
- Langfermann, D.S.; Rössler, O.G.; Thiel, G. Stimulation of B-Raf increases c-Jun and c-Fos expression and upregulates AP-1-regulated gene transcription in insulinoma cells. Mol. Cell. Endocrinol. 2018, 472, 126–139. [Google Scholar] [CrossRef]
- Thiel, G.; Müller, I.; Rössler, O.G. Signal transduction via TRPM3 in pancreatic β-cells. J. Mol. Endocrinol. 2013, 50, R75–R83. [Google Scholar] [CrossRef] [Green Version]
- Thiel, G.; Rubil, S.; Lesch, A.; Guethlein, L.A.; Rössler, O.G. Transient receptor potential TRPM3 channels: Pharmacology, signaling, and biological functions. Pharmacol. Res. 2017, 124, 92–99. [Google Scholar] [CrossRef]
- Rubil, S.; Rössler, O.G.; Thiel, G. CREB, AP-1, ternary complex factors and MAP kinases connect transient receptor potential melastatin-3 (TRPM3) channel stimulation with increased c-Fos expression. Br. J. Pharmacol. 2016, 173, 305–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.S.; Treisman, R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995, 14, 5037–5047. [Google Scholar] [CrossRef] [PubMed]
- Ely, H.A.; Mellon, P.L.; Coss, D. GnRH induces the c-Fos gene via phosphorylation of SRF by the calcium/calmodulin kinase II pathway. Mol. Endocrinol. 2011, 25, 669–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glidewell-Kenney, C.A.; Trang, C.; Shao, P.P.; Gutierrez-Reed, N.; Uzo-Okereke, A.M.; Coss, D.; Mellon, P.L. Neurokinin B induces c-fos transcription via protein kinase C and activation of serum response factor and Elk-1 in immortalized GnRH neurons. Endocrinology 2014, 155, 3909–3919. [Google Scholar] [CrossRef] [Green Version]
- Shore, P.; Sharrocks, A.D. The transcription factor Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 1994, 14, 3283–3291. [Google Scholar] [CrossRef] [Green Version]
- Ling, Y.; Lakey, J.H.; Roberts, C.E.; Sharrocks, A.D. Molecular characterization of the B-box protein-protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J. 1997, 16, 2431–2440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassler, M.; Richmond, T.J. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 2001, 20, 3013–3028. [Google Scholar] [CrossRef] [Green Version]
- Zeke, A.; Misheva, M.; Reményi, A.; Bogoyevitch, M.A. JNK signaling regulation and functions based on complex protein-protein partnerships. Microbiol. Mol. Biol. Rev. 2016, 80, 793–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavoie, H.; Gagnon, J.; Therrien, M. ERK signalling: A master regulator of cell behaviour, life, and fate. Nat. Rev. Mol. Cell Biol. 2020, 21, 607–632. [Google Scholar] [CrossRef] [PubMed]
- Canovas, B.; Nebreda, A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Ducret, C.; Maira, S.-M.; Lutz, Y.; Wasylyk, B. The ternary complex factor. Net contains two distinct elements that mediate different responses to MAP kinase signalling cascades. Oncogene 2000, 19, 5063–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mylona, A.; Theillet, F.-X.; Foster, C.; Cheng, T.M.; Mirralles, F.; Bates, P.A.; Selenko, P.; Treisman, R. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science 2016, 354, 233–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strahl, T.; Gille, H.; Shaw, P.E. Selective response of ternary complex factor SAP1a to different mitogen-activated protein kinase subgroups. Proc. Natl. Acad. Sci. USA 1996, 93, 11563–11568. [Google Scholar] [CrossRef] [Green Version]
- Janknecht, R.; Hunter, T. Activation of the Sap-1a transcription factor by the c-Jun N-terminal kinase (JNK) mitogen-activated protein kinase. J. Biol. Chem. 1997, 272, 4219–4224. [Google Scholar] [CrossRef] [Green Version]
- Janknecht, R.; Hunter, T. Convergence of MAP kinase pathways in the ternary complex factor Sap-1a. EMBO J. 1997, 16, 1620–1627. [Google Scholar] [CrossRef] [Green Version]
- Raingeaud, J.; Whitmarsh, A.J.; Barrett, T.; Dérijard, B.; Davis, R.J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signaling pathway. Mol. Cell. Biol. 1996, 16, 1247–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.-H.; Yates, P.R.; Whitmarsh, A.J.; Davis, R.J.; Sharrocks, A.D. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting domain. Mol. Cell. Biol. 1998, 18, 710–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.-H.; Whitmarsh, A.J.; Davis, R.J.; Sharrocks, A.D. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998, 17, 1740–1749. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, D.; Glossip, D.; Xing, H.; Muslin, A.J.; Kornfeld, K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999, 13, 163–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheridan, D.L.; Kong, Y.; Parker, S.A.; Dalby, K.N.; Turk, B.E. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J. Biol. Chem. 2008, 283, 19511–19520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantz, D.A.; Jacobs, D.; Glossip, D.; Kornfeld, K. Docking sites on substrate proteins direct extracellular signal-regulated kinase to phosphorylate specific residues. J. Biol. Chem. 2001, 276, 27256–27265. [Google Scholar] [CrossRef] [Green Version]
- Stevens, J.L.; Cantin, G.T.; Wang, G.; Shevchenko, A.; Shevchenko, A.; Berk, A.J. Transcriptional control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 2002, 269, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Balamotis, M.A.; Stevens, J.L.; Yamaguchi, Y.; Handa, H.; Berk, A.J. Mediator recruitment for both recruitment and postrecruitment steps in transcriptional initiation. Mol. Cell 2005, 17, 683–694. [Google Scholar] [CrossRef]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.; Goodman, R.H. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001, 276, 13505–13508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.-J.; Yang, S.-H.; Maeda, Y.; Sladek, F.M.; Sharrocks, A.D.; Martins-Green, M. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 2003, 22, 281–291. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, T.; Stewart, S.; Guan, K.-L. The calcium/calmodulin-dependent protein phosphatase calcineurin is the major Elk-1 phosphatase. J. Biol. Chem. 1997, 272, 29415–29418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Karin, M. Stimulation of Elk1 transcriptional activity by mitogen-activated protein kinases is negatively regulated by protein phosphatase 2B (calcineurin). J. Biol. Chem. 1999, 274, 15173–15180. [Google Scholar] [CrossRef] [Green Version]
- Thiel, G.; Schmidt, T.; Rössler, O.G. Ca2+ microdomains, calcineurin, and the regulation of gene transcription. Cells 2021, 10, 875. [Google Scholar] [CrossRef]
- Langfermann, D.S.; Schmidt, T.; Rössler, O.G.; Thiel, G. Calcineurin controls gene transcription following stimulation of a Gαq-coupled designer receptor. Exp. Cell Res. 2019, 383, 111553. [Google Scholar] [CrossRef]
- Lam, B.Y.H.; Zhang, W.; Enticknap, N.; Haggis, E.; Cader, M.Z.; Chawla, S. Inverse regulation of plasticity-related immediate early genes by calcineurin in hippocampal neurons. J. Biol. Chem. 2009, 284, 12562–12571. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.R.; Rao, A. NFAT, immunity and cancer: A transcription factor comes of age. Nat. Rev. Immunol. 2010, 10, 645–656. [Google Scholar] [CrossRef]
- Yang, S.-H.; Vickers, E.; Brehm, A.; Kouzarides, T.; Sharrocks, A.D. Temporal recruitment of the mSin3A-histone deacetylase complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 2001, 21, 2802–2814. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-H.; Bumpass, D.C.; Perkins, N.D.; Sharrocks, A.D. The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol. Cell. Biol. 2002, 22, 5036–5046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.-H.; Jaffray, E.; Hay, R.T.; Sharrocks, A.D. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 2003, 12, 63–74. [Google Scholar] [CrossRef]
- Yang, S.-H.; Sharrocks, A.D. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 2004, 13, 611–617. [Google Scholar] [CrossRef]
- Yang, S.-H.; Galanis, A.; Witty, J.; Sharrocks, A.D. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006, 25, 5083–5093. [Google Scholar] [CrossRef] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiel, G.; Backes, T.M.; Guethlein, L.A.; Rössler, O.G. Critical Protein–Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription. Molecules 2021, 26, 6125. https://doi.org/10.3390/molecules26206125
Thiel G, Backes TM, Guethlein LA, Rössler OG. Critical Protein–Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription. Molecules. 2021; 26(20):6125. https://doi.org/10.3390/molecules26206125
Chicago/Turabian StyleThiel, Gerald, Tobias M. Backes, Lisbeth A. Guethlein, and Oliver G. Rössler. 2021. "Critical Protein–Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription" Molecules 26, no. 20: 6125. https://doi.org/10.3390/molecules26206125