Interrogating Plant-Microbe Interactions with Chemical Tools: Click Chemistry Reagents for Metabolic Labeling and Activity-Based Probes
Abstract
:1. Introduction
2. Chemical Biology Toolbox
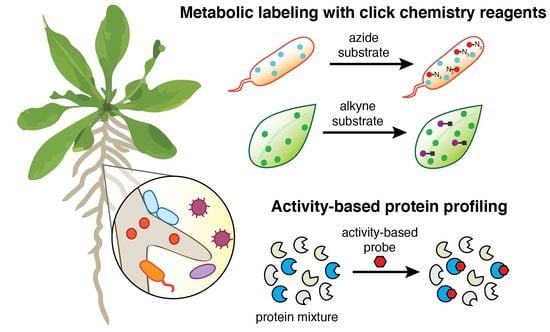
2.1. Bioorthogonal Click Chemistry Reagents for Metabolic Labeling
2.2. Activity-Based Probes for Functionally Profiling Plant-Microbe Systems
3. Challenges for Chemical Biology in Plant-Microbe Interactions
3.1. Applying Chemical Biology Tools in Laboratory Models vs. Field Systems
3.2. Delivering Chemical Tools to Live Plant Tissues
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Sadler, N.C.; Wright, A.T. Activity-based protein profiling of microbes. Curr. Opin. Chem. Biol. 2015, 24, 139–144. [Google Scholar] [CrossRef] [Green Version]
- Whidbey, C.; Sadler, N.C.; Nair, R.N.; Volk, R.F.; DeLeon, A.J.; Bramer, L.M.; Fansler, S.J.; Hansen, J.R.; Shukla, A.K.; Jansson, J.K.; et al. A Probe-Enabled Approach for the Selective Isolation and Characterization of Functionally Active Subpopulations in the Gut Microbiome. J. Am. Chem. Soc. 2019, 141, 42–47. [Google Scholar] [CrossRef]
- Koppel, N.; Balskus, E.P. Exploring and Understanding the Biochemical Diversity of the Human Microbiota. Cell Chem. Biol. 2016, 23, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlechter, R.O.; Miebach, M.; Remus-Emsermann, M.N.P. Driving factors of epiphytic bacterial communities: A review. J. Adv. Res. 2019, 19, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Vincent, D.; Rafiqi, M.; Job, D. The Multiple Facets of Plant-Fungal Interactions Revealed through Plant and Fungal Secretomics. Front. Plant Sci. 2019, 10, 1626. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Herve, V.; Labbe, J.; Lastovetsky, O.A.; et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [Green Version]
- O&Banion, B.S.; O&Neal, L.; Alexandre, G.; Lebeis, S.L. Bridging the Gap Between Single-Strain and Community-Level Plant-Microbe Chemical Interactions. Mol. Plant-Microbe Interact. 2020, 33, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Ke, P.J.; Wan, J. Effects of soil microbes on plant competition: A perspective from modern coexistence theory. Ecol. Monogr. 2019, 90, e01391. [Google Scholar] [CrossRef]
- Chagas, F.O.; Pessotti, R.C.; Caraballo-Rodriguez, A.M.; Pupo, M.T. Chemical signaling involved in plant-microbe interactions. Chem. Soc. Rev. 2018, 47, 1652–1704. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The Chemistry of Plant-Microbe Interactions in the Rhizosphere and the Potential for Metabolomics to Reveal Signaling Related to Defense Priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [Green Version]
- Gregor, R.; David, S.; Meijler, M.M. Chemical strategies to unravel bacterial-eukaryotic signaling. Chem. Soc. Rev. 2018, 47, 1761–1772. [Google Scholar] [CrossRef]
- Mazzoni-Putman, S.M.; Stepanova, A.N. A Plant Biologist’s Toolbox to Study Translation. Front. Plant Sci. 2018, 9, 873. [Google Scholar] [CrossRef]
- Van der Hoorn, R.A.; Colby, T.; Nickel, S.; Richau, K.H.; Schmidt, J.; Kaiser, M. Mining the Active Proteome of Arabidopsis thaliana. Front. Plant Sci. 2011, 2, 89. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekar, B.; Colby, T.; Emran Khan Emon, A.; Jiang, J.; Hong, T.N.; Villamor, J.G.; Harzen, A.; Overkleeft, H.S.; van der Hoorn, R.A. Broad-range glycosidase activity profiling. Mol. Cell Proteom. 2014, 13, 2787–2800. [Google Scholar] [CrossRef] [Green Version]
- Row, R.D.; Prescher, J.A. Constructing New Bioorthogonal Reagents and Reactions. ACC Chem. Res. 2018, 51, 1073–1081. [Google Scholar] [CrossRef]
- McKay, C.S.; Finn, M.G. Click chemistry in complex mixtures: Bioorthogonal bioconjugation. Chem. Biol. 2014, 21, 1075–1101. [Google Scholar] [CrossRef] [Green Version]
- Devaraj, N.K. The Future of Bioorthogonal Chemistry. ACS Cent. Sci. 2018, 4, 952–959. [Google Scholar] [CrossRef] [Green Version]
- Agard, N.J.; Baskin, J.M.; Prescher, J.A.; Lo, A.; Bertozzi, C.R. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006, 1, 644–648. [Google Scholar] [CrossRef]
- Bukowski, N.; Pandey, J.L.; Doyle, L.; Richard, T.L.; Anderson, C.T.; Zhu, Y. Development of a clickable designer monolignol for interrogation of lignification in plant cell walls. Bioconjug. Chem. 2014, 25, 2189–2196. [Google Scholar] [CrossRef]
- Pandey, J.L.; Kiemle, S.N.; Richard, T.L.; Zhu, Y.; Cosgrove, D.J.; Anderson, C.T. Investigating Biochemical and Developmental Dependencies of Lignification with a Click-Compatible Monolignol Analog in Arabidopsis thaliana Stems. Front. Plant Sci. 2016, 7, 1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, J.L.; Wang, B.; Diehl, B.G.; Richard, T.L.; Chen, G.; Anderson, C.T. A versatile click-compatible monolignol probe to study lignin deposition in plant cell walls. PLoS ONE 2015, 10, e0121334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, C.; Lion, C.; Huss, B.; Blervacq, A.S.; Spriet, C.; Guerardel, Y.; Biot, C.; Hawkins, S. BLISS: Shining a light on lignification in plants. Plant Signal. Behav. 2017, 12, e1359366. [Google Scholar] [CrossRef] [PubMed]
- Hoogenboom, J.; Berghuis, N.; Cramer, D.; Geurts, R.; Zuilhof, H.; Wennekes, T. Direct imaging of glycans in Arabidopsis roots via click labeling of metabolically incorporated azido-monosaccharides. BMC Plant Biol. 2016, 16, 220. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.T.; Wallace, I.S.; Somerville, C.R. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc. Natl. Acad. Sci. USA 2012, 109, 1329–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumont, M.; Lehner, A.; Vauzeilles, B.; Malassis, J.; Marchant, A.; Smyth, K.; Linclau, B.; Baron, A.; Mas Pons, J.; Anderson, C.T.; et al. Plant cell wall imaging by metabolic click-mediated labelling of rhamnogalacturonan II using azido 3-deoxy-D-manno-oct-2-ulosonic acid. Plant J. 2016, 85, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Glenn, W.S.; Stone, S.E.; Ho, S.H.; Sweredoski, M.J.; Moradian, A.; Hess, S.; Bailey-Serres, J.; Tirrell, D.A. Bioorthogonal Noncanonical Amino Acid Tagging (BONCAT) Enables Time-Resolved Analysis of Protein Synthesis in Native Plant Tissue. Plant Physiol. 2017, 173, 1543–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatzenpichler, R.; Scheller, S.; Tavormina, P.L.; Babin, B.M.; Tirrell, D.A.; Orphan, V.J. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ. Microbiol. 2014, 16, 2568–2590. [Google Scholar] [CrossRef]
- Steward, K.F.; Eilers, B.; Tripet, B.; Fuchs, A.; Dorle, M.; Rawle, R.; Soriano, B.; Balasubramanian, N.; Copie, V.; Bothner, B.; et al. Metabolic Implications of Using BioOrthogonal Non-Canonical Amino Acid Tagging (BONCAT) for Tracking Protein Synthesis. Front. Microbiol. 2020, 11, 197. [Google Scholar] [CrossRef]
- Boyle, P.C.; Schwizer, S.; Hind, S.R.; Kraus, C.M.; De la Torre Diaz, S.; He, B.; Martin, G.B. Detecting N-myristoylation and S-acylation of host and pathogen proteins in plants using click chemistry. Plant Methods 2016, 12, 38. [Google Scholar] [CrossRef] [Green Version]
- Perez, A.J.; Bode, H.B. “Click Chemistry” for the Simple Determination of Fatty-Acid Uptake and Degradation: Revising the Role of Fatty-Acid Transporters. ChemBioChem 2015, 16, 1588–1591. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.J.; Bode, H.B. ω-Azido fatty acids as probes to detect fatty acid biosynthesis, degradation, and modification. J. Lipid Res. 2014, 55, 1897–1901. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Petti, C.; Williams, M.A.; DeBolt, S. Experimental approaches to study plant cell walls during plant-microbe interactions. Front. Plant Sci. 2014, 5, 540. [Google Scholar] [CrossRef] [Green Version]
- Paper, J.M.; Mukherjee, T.; Schrick, K. Bioorthogonal click chemistry for fluorescence imaging of choline phospholipids in plants. Plant Methods 2018, 14, 31. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, K.; van der Hoorn, R.A. The Increasing Impact of Activity-Based Protein Profiling in Plant Science. Plant Cell Physiol. 2016, 57, 446–461. [Google Scholar] [CrossRef] [Green Version]
- Hunerdosse, D.; Nomura, D.K. Activity-based proteomic and metabolomic approaches for understanding metabolism. Curr. Opin. Biotechnol. 2014, 28, 116–126. [Google Scholar] [CrossRef] [Green Version]
- Whidbey, C.; Wright, A.T. Activity-Based Protein Profiling-Enabling Multimodal Functional Studies of Microbial Communities. In Activity-Based Protein Profiling; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 420, pp. 1–21. [Google Scholar]
- Bolger, M.E.; Arsova, B.; Usadel, B. Plant genome and transcriptome annotations: From misconceptions to simple solutions. Brief. Bioinform. 2018, 19, 437–449. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Yang, F.; Stepanauskas, R.; Cardenas, E.; Garoutte, A.; Williams, R.; Flater, J.; Tiedje, J.M.; Hofmockel, K.S.; Gelder, B.; et al. Strategies to improve reference databases for soil microbiomes. ISME J. 2017, 11, 829–834. [Google Scholar] [CrossRef] [Green Version]
- Bharti, R.; Grimm, D.G. Current challenges and best-practice protocols for microbiome analysis. Brief. Bioinform. 2019, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome-from metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef] [Green Version]
- Misas-Villamil, J.C.; Toenges, G.; Kolodziejek, I.; Sadaghiani, A.M.; Kaschani, F.; Colby, T.; Bogyo, M.; van der Hoorn, R.A. Activity profiling of vacuolar processing enzymes reveals a role for VPE during oomycete infection. Plant J. 2013, 73, 689–700. [Google Scholar] [CrossRef]
- Buscaill, P.; Chandrasekar, B.; Sanguankiattichai, N.; Kourelis, J.; Kaschani, F.; Thomas, E.L.; Morimoto, K.; Kaiser, M.; Preston, G.M.; Ichinose, Y.; et al. Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 2019, 364, 6436. [Google Scholar] [CrossRef]
- Paulus, J.K.; Kourelis, J.; Ramasubramanian, S.; Homma, F.; Godson, A.; Horger, A.C.; Hong, T.N.; Krahn, D.; Ossorio Carballo, L.; Wang, S.; et al. Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc. Natl. Acad. Sci. USA 2020, 117, 17409–17417. [Google Scholar] [CrossRef] [PubMed]
- Franco, J.; Thapa, S.P.; Pang, Z.; Gurung, F.B.; Liebrand, T.W.; Stevens, D.M.; Ancona, V.; Wang, N.; Coaker, G. Citrus vascular proteomics highlights the role of peroxidases and serine proteases during Huanglongbing disease progression. Mol. Cell Proteom. 2020, 19, 1936–1951. [Google Scholar] [CrossRef]
- Clark, K.; Franco, J.Y.; Schwizer, S.; Pang, Z.; Hawara, E.; Liebrand, T.W.H.; Pagliaccia, D.; Zeng, L.; Gurung, F.B.; Wang, P.; et al. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 2018, 9, 1718. [Google Scholar] [CrossRef]
- Pogorelko, G.V.; Juvale, P.S.; Rutter, W.B.; Hutten, M.; Maier, T.R.; Hewezi, T.; Paulus, J.; van der Hoorn, R.A.; Grundler, F.M.; Siddique, S.; et al. Re-targeting of a plant defense protease by a cyst nematode effector. Plant J. 2019, 98, 1000–1014. [Google Scholar] [CrossRef] [Green Version]
- Hutten, M.; Geukes, M.; Misas-Villamil, J.C.; van der Hoorn, R.A.; Grundler, F.M.; Siddique, S. Activity profiling reveals changes in the diversity and activity of proteins in Arabidopsis roots in response to nematode infection. Plant Physiol. Biochem. 2015, 97, 36–43. [Google Scholar] [CrossRef]
- Planas-Marques, M.; Bernardo-Faura, M.; Paulus, J.; Kaschani, F.; Kaiser, M.; Valls, M.; van der Hoorn, R.A.L.; Coll, N.S. Protease Activities Triggered by Ralstonia solanacearum Infection in Susceptible and Tolerant Tomato Lines. Mol. Cell Proteom. 2018, 17, 1112–1125. [Google Scholar] [CrossRef] [Green Version]
- Schroder, S.P.; de Boer, C.; McGregor, N.G.S.; Rowland, R.J.; Moroz, O.; Blagova, E.; Reijngoud, J.; Arentshorst, M.; Osborn, D.; Morant, M.D.; et al. Dynamic and Functional Profiling of Xylan-Degrading Enzymes in Aspergillus Secretomes Using Activity-Based Probes. ACS Cent. Sci. 2019, 5, 1067–1078. [Google Scholar] [CrossRef] [Green Version]
- Kaschani, F.; Gu, C.; van der Hoorn, R.A. Activity-based protein profiling of infected plants. Methods Mol. Biol. 2012, 835, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Shabab, M.; Shindo, T.; Gu, C.; Kaschani, F.; Pansuriya, T.; Chintha, R.; Harzen, A.; Colby, T.; Kamoun, S.; van der Hoorn, R.A. Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 2008, 20, 1169–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berns, A.E.; Philipp, H.; Narres, H.D.; Burauel, P.; Vereecken, H.; Tappe, W. Effect of gamma-sterilization and autoclaving on soil organic matter structure as studied by solid state NMR, UV and fluorescence spectroscopy. Eur. J. Soil Sci. 2008, 59, 540–550. [Google Scholar] [CrossRef]
- Calvet, R. Adsorption of organic chemicals in soils. Environ. Health Perspect. 1989, 83, 145–177. [Google Scholar] [CrossRef]
- Mamy, L.; Patureau, D.; Barriuso, E.; Bedos, C.; Bessac, F.; Louchart, X.; Martin-Laurent, F.; Miege, C.; Benoit, P. Prediction of the Fate of Organic Compounds in the Environment From Their Molecular Properties: A Review. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1277–1377. [Google Scholar] [CrossRef] [Green Version]
- Pierret, A.; Moran, C.J. Plant Roots and Soil Structure. In Encyclopedia of Agrophysics; Gliński, J., Horabik, J., Lipiec, J., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 628–632. [Google Scholar] [CrossRef]
- Bach, E.M.; Williams, R.J.; Hargreaves, S.K.; Yang, F.; Hofmockel, K.S. Greatest soil microbial diversity found in micro-habitats. Soil Biol. Biochem. 2018, 118, 217–226. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Kopriva, S. Metabolic niches in the rhizosphere microbiome: New tools and approaches to analyse metabolic mechanisms of plant-microbe nutrient exchange. J. Exp. Bot. 2019, 70, 1087–1094. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Pronk, G.J.; Heister, K.; Kogel-Knabner, I.; Martens, R.; Tebbe, C.C. Artificial soil studies reveal domain-specific preferences of microorganisms for the colonisation of different soil minerals and particle size fractions. FEMS Microbiol. Ecol. 2014, 90, 770–782. [Google Scholar] [CrossRef]
- Fichman, Y.; Miller, G.; Mittler, R. Whole-Plant Live Imaging of Reactive Oxygen Species. Mol. Plant 2019, 12, 1203–1210. [Google Scholar] [CrossRef] [Green Version]
- Thomanek, H.; Schenk, S.T.; Stein, E.; Kogel, K.H.; Schikora, A.; Maison, W. Modified N-acyl-homoserine lactones as chemical probes for the elucidation of plant-microbe interactions. Org. Biomol. Chem. 2013, 11, 6994–7003. [Google Scholar] [CrossRef]
- Mravec, J.; Kracun, S.K.; Zemlyanskaya, E.; Rydahl, M.G.; Guo, X.; Picmanova, M.; Sorensen, K.K.; Ruzicka, K.; Willats, W.G.T. Click chemistry-based tracking reveals putative cell wall-located auxin binding sites in expanding cells. Sci. Rep. 2017, 7, 15988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaschani, F.; Verhelst, S.H.; van Swieten, P.F.; Verdoes, M.; Wong, C.S.; Wang, Z.; Kaiser, M.; Overkleeft, H.S.; Bogyo, M.; van der Hoorn, R.A. Minitags for small molecules: Detecting targets of reactive small molecules in living plant tissues using ‘click chemistry’. Plant J. 2009, 57, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems Biology of Plant-Microbiome Interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dejonghe, W.; Okamoto, M.; Cutler, S.R. Small Molecule Probes of ABA Biosynthesis and Signaling. Plant Cell Physiol. 2018, 59, 1490–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meesters, C.; Monig, T.; Oeljeklaus, J.; Krahn, D.; Westfall, C.S.; Hause, B.; Jez, J.M.; Kaiser, M.; Kombrink, E. A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat. Chem. Biol. 2014, 10, 830–836. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, V.S. Interrogating Plant-Microbe Interactions with Chemical Tools: Click Chemistry Reagents for Metabolic Labeling and Activity-Based Probes. Molecules 2021, 26, 243. https://doi.org/10.3390/molecules26010243
Lin VS. Interrogating Plant-Microbe Interactions with Chemical Tools: Click Chemistry Reagents for Metabolic Labeling and Activity-Based Probes. Molecules. 2021; 26(1):243. https://doi.org/10.3390/molecules26010243
Chicago/Turabian StyleLin, Vivian S. 2021. "Interrogating Plant-Microbe Interactions with Chemical Tools: Click Chemistry Reagents for Metabolic Labeling and Activity-Based Probes" Molecules 26, no. 1: 243. https://doi.org/10.3390/molecules26010243