Cyclic RGD and isoDGR Integrin Ligands Containing cis-2-amino-1-cyclopentanecarboxylic (cis-β-ACPC) Scaffolds
Abstract
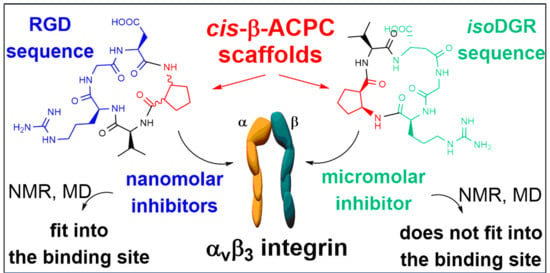
:1. Introduction
2. Results
2.1. Synthesis
2.2. Biological Assays
2.3. NMR Studies
2.3.1. Conformational Analysis of Compound 8
2.3.2. Conformational Analysis of Compound 9
2.3.3. Conformational Analysis of Compound 10
2.4. Computational Studies
3. Material and Methods
3.1. General Procedures
3.2. Competitive Binding Assays to the Purified αvβ3 and α5β1 Receptors
3.3. Cell Adhesion Assays
3.4. NMR Studies
3.5. Computational Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef] [Green Version]
- Takada, Y.; Ye, X.; Simon, S. The Integrins. Genome Biol. 2007, 8, 215. [Google Scholar] [CrossRef] [Green Version]
- Avraamides, C.J.; Garmy-Susini, B.; Varner, J.A. Integrins in angiogenesis and lymphangiogenesis. Nat. Rev. Cancer 2008, 8, 604–617. [Google Scholar] [CrossRef] [Green Version]
- Danhier, F.; Le Breton, A.; Préat, V. RGD-based strategies to target αVβ3 integrin in cancer therapy and diagnosis. Mol. Pharm. 2012, 9, 2961–2973. [Google Scholar] [CrossRef] [PubMed]
- Nieberler, M.; Reuning, U.; Reichart, F.; Notni, J.; Wester, H.-J.; Schwaiger, M.; Weinmüller, M.; Räder, A.; Steiger, K.; Kessler, H. Exploring the role of RGD-recognizing integrins in cancer. Cancers 2017, 9, 116. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef]
- Spitaleri, A.; Mari, S.; Curnis, F.; Traversari, C.; Longhi, R.; Bordignon, C.; Corti, A.; Rizzardi, G.-P.; Musco, G. Structural basis for the interaction of isoDGR with the RGD-binding site of αVβ3 integrin. J. Biol. Chem. 2008, 283, 19757–19768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curnis, F.; Cattaneo, A.; Longhi, R.; Sacchi, A.; Gasparri, A.M.; Pastorino, F.; Di Matteo, P.; Traversari, C.; Bachi, A.; Ponzoni, M.; et al. Critical role of flanking residues in NGR-to-isoDGR transition and CD13/integrin receptor switching. J. Biol. Chem. 2010, 285, 9114–9123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghitti, M.; Spitaleri, A.; Valentinis, B.; Mari, S.; Asperti, C.; Traversari, C.; Rizzardi, G.-P.; Musco, G. Molecular dynamics reveal that isoDGR-containing cyclopeptides are true αVβ3 antagonists unable to promote integrin allostery and activation. Angew. Chem. Int. Ed. 2012, 51, 7702–7705. [Google Scholar] [CrossRef] [PubMed]
- Paladino, A.; Civera, M.; Curnis, F.; Paolillo, M.; Gennari, C.; Piarulli, U.; Corti, A.; Belvisi, L.; Colombo, G. The Importance of Detail: How Differences in Ligand Structures Determine Distinct Functional Responses in Integrin αVβ3. Chem. Eur. J. 2019, 25, 5959–5970. [Google Scholar] [CrossRef]
- Aumailley, M.; Gurrath, M.; Müller, G.; Calvete, J.; Timpl, R.; Kessler, H. Arg-Gly-Asp constrained within cyclic pentapoptides Strong and selective inhibitors of cell adhesion to vitronectin and laminin fragment P1. FEBS Lett. 1991, 291, 50–54. [Google Scholar] [CrossRef] [Green Version]
- Auzzas, L.; Zanardi, F.; Battistini, L.; Burreddu, P.; Carta, P.; Rassu, G.; Curti, C.; Casiraghi, G. Targeting αVβ3 integrin: Design and applications of mono- and multifunctional RGD-based peptides and semipeptides. Curr. Med. Chem. 2010, 17, 1255–1299. [Google Scholar] [CrossRef] [PubMed]
- Hatley, R.J.D.; Macdonald, S.J.F.; Slack, R.J.; Le, J.; Ludbrook, S.B.; Lukey, P.T. An αv-RGD Integrin Inhibitor Toolbox: Drug Discovery Insight, Challenges and Opportunities. Angew. Chem. Int. Ed. 2018, 57, 3298–3321. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, R.; Schembri, L.; Piarulli, U.; Pinoli, M.; Rasini, E.; Paolillo, M.; Galiazzo, M.C.; Cosentino, M.; Marino, F. Effects of a novel cyclic RGD peptidomimetic on cell proliferation, migration and angiogenic activity in human endothelial cells. Vasc. Cell 2014, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, R.; Mingozzi, M.; Belvisi, L.; Arosio, D.; Piarulli, U.; Carenini, N.; Perego, P.; Zaffaroni, N.; De Cesare, M.; Castiglioni, V.; et al. Synthesis and biological evaluation (in Vitro and in Vivo) of cyclic arginine-glycine-aspartate (RGD) peptidomimetic-paclitaxel conjugates targeting integrin αVβ3. J. Med. Chem. 2012, 55, 10460–10474. [Google Scholar] [CrossRef] [Green Version]
- Dal Corso, A.; Caruso, M.; Belvisi, L.; Arosio, D.; Piarulli, U.; Albanese, C.; Gasparri, F.; Marsiglio, A.; Sola, F.; Troiani, S.; et al. Synthesis and biological evaluation of RGD peptidomimetic-paclitaxel conjugates bearing lysosomally cleavable linkers. Chem. Eur. J. 2015, 21, 6921–6929. [Google Scholar] [CrossRef]
- Zanella, S.; Angerani, S.; Pina, A.; López Rivas, P.; Giannini, C.; Panzeri, S.; Arosio, D.; Caruso, M.; Gasparri, F.; Fraietta, I.; et al. Tumor Targeting with an isoDGR–Drug Conjugate. Chem. Eur. J. 2017, 23, 7910–7914. [Google Scholar] [CrossRef] [Green Version]
- López Rivas, P.; Bodero, L.; Korsak, B.; Hechler, T.; Pahl, A.; Müller, C.; Arosio, D.; Pignataro, L.; Gennari, C.; Piarulli, U. Synthesis and biological evaluation of RGD and isoDGR peptidomimetic-α-amanitin conjugates for tumor-targeting. Beilstein J. Org. Chem. 2018, 14, 407–415. [Google Scholar]
- Raposo Moreira Dias, A.; Bodero, L.; Martins, A.; Arosio, D.; Gazzola, S.; Belvisi, L.; Pignataro, L.; Steinkühler, C.; Dal Corso, A.; Gennari, C.; et al. Synthesis and Biological Evaluation of RGD and isoDGR–Monomethyl Auristatin Conjugates Targeting Integrin αVβ3. ChemMedChem 2019, 14, 938–942. [Google Scholar] [CrossRef] [Green Version]
- Feni, L.; Parente, S.; Robert, C.; Gazzola, S.; Arosio, D.; Piarulli, U.; Neundorf, I. Kiss and run: Promoting effective and targeted cellular uptake of a drug delivery vehicle composed of an integrin-targeting diketopiperazine peptidomimetic and a cell-penetrating peptide. Bioconjugate Chem. 2019, 307, 2011–2022. [Google Scholar] [CrossRef] [Green Version]
- Agnello, S.; Brand, M.; Chellat, M.F.; Gazzola, S.; Riedl, R. A Structural View on Medicinal Chemistry Strategies against Drug Resistance. Angew. Chem. Int. Ed. 2019, 58, 3300–3345. [Google Scholar] [CrossRef] [PubMed]
- Dechantsreiter, M.A.; Planker, E.; Mathä, B.; Lohof, E.; Hölzemann, G.; Jonczyk, A.; Goodman, S.L.; Kessler, H. N-methylated cyclic RGD peptides as highly active and selective αVβ3 integrin antagonists. J. Med. Chem. 1999, 42, 3033–3040. [Google Scholar] [CrossRef] [PubMed]
- Belvisi, L.; Riccioni, T.; Marcellini, M.; Vesci, L.; Chiarucci, I.; Efrati, D.; Potenza, D.; Scolastico, C.; Manzoni, L.; Lombardo, K.; et al. Biological and molecular properties of a new αVβ3/αVβ5 integrin antagonist. Mol. Cancer Ther. 2005, 4, 1670–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belvisi, L.; Bernardi, A.; Colombo, M.; Manzoni, L.; Potenza, D.; Scolastico, G.; Giannini, G.; Marcellini, M.; Riccioni, T.; Castorina, M.; et al. Targeting integrins: Insights into structure and activity of cyclic RGD pentapeptide mimics containing azabicycloalkane amino acids. Bioorg. Med. Chem. 2006, 14, 169–180. [Google Scholar] [CrossRef]
- Manzoni, L.; Belvisi, L.; Arosio, D.; Civera, M.; Pilkington-Miksa, M.; Potenza, D.; Caprini, A.; Araldi, E.M.V.; Monferrini, E.; Mancino, M.; et al. Cyclic RGD-Containing functionalized azabicycloalkane peptides as potent integrin antagonists for tumor targeting. ChemMedChem 2009, 4, 615–632. [Google Scholar] [CrossRef]
- Zanardi, F.; Burreddu, P.; Rassu, G.; Auzzas, L.; Battistini, L.; Curti, C.; Sartori, A.; Nicastro, G.; Menchi, G.; Cini, N.; et al. Discovery of subnanomolar arginine-glycine-aspartate-based αVβ3/αVβ5 integrin binders embedding 4-aminoproline residues. J. Med. Chem. 2008, 51, 1771–1782. [Google Scholar] [CrossRef]
- Ressurreiçao, A.S.M.; Vidu, A.; Civera, M.; Belvisi, L.; Potenza, D.; Manzoni, L.; Ongeri, S.; Gennari, C.; Piarulli, U. Cyclic RGD-peptidomimetics containing bifunctional diketopiperazine scaffolds as new potent integrin ligands. Chem. Eur. J. 2009, 15, 12184–12188. [Google Scholar] [CrossRef]
- Marchini, M.; Mingozzi, M.; Colombo, R.; Guzzetti, I.; Belvisi, L.; Vasile, F.; Potenza, D.; Piarulli, U.; Arosio, D.; Gennari, C. Cyclic RGD peptidomimetics containing bifunctional diketopiperazine scaffolds as new potent integrin ligands. Chem. Eur. J. 2012, 18, 6195–6207. [Google Scholar] [CrossRef] [Green Version]
- Mingozzi, M.; Dal Corso, A.; Marchini, M.; Guzzetti, I.; Civera, M.; Piarulli, U.; Arosio, D.; Belvisi, L.; Potenza, D.; Pignataro, L.; et al. Cyclic isoDGR peptidomimetics as low-nanomolar αVβ3 integrin ligands. Chem. Eur. J. 2013, 19, 3563–3567. [Google Scholar] [CrossRef]
- Panzeri, S.; Zanella, S.; Arosio, D.; Vahdati, L.; Dal Corso, A.; Pignataro, L.; Paolillo, M.; Schinelli, S.; Belvisi, L.; Gennari, C.; et al. Cyclic isoDGR and RGD Peptidomimetics Containing Bifunctional Diketopiperazine Scaffolds are Integrin Antagonists. Chem. Eur. J. 2015, 21, 6265–6271. [Google Scholar] [CrossRef]
- Nardelli, F.; Paissoni, C.; Quilici, G.; Gori, A.; Traversari, C.; Valentinis, B.; Sacchi, A.; Corti, A.; Curnis, F.; Ghitti, M.; et al. Succinimide-Based Conjugates Improve IsoDGR Cyclopeptide Affinity to αVβ3 without Promoting Integrin Allosteric Activation. J. Med. Chem. 2018, 61, 7474–7485. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.O.; Otto, E.; Mas-Moruno, C.; Schiller, H.B.; Marinelli, L.; Cosconati, S.; Bochen, A.; Vossmeyer, D.; Zahn, G.; Stragies, R.; et al. Conformational control of integrin-subtype selectivity in isoDGR peptide motifs: A biological switch. Angew. Chem. Int. Ed. 2010, 49, 9278–9281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrele, C.; Martinek, T.A.; Reiser, O.; Berlicki, L. Peptides containing β-amino acid patterns: Challenges and successes in medicinal chemistry. J. Med. Chem. 2014, 57, 9718–9739. [Google Scholar] [CrossRef] [PubMed]
- Gentilucci, L.; Gallo, F.; Meloni, F.; Mastandrea, M.; Del Secco, B.; De Marco, R. Controlling Cyclopeptide Backbone Conformation with β/α-Hybrid Peptide–Heterocycle Scaffolds. Eur. J. Org. Chem. 2016, 2016, 3243–3251. [Google Scholar] [CrossRef]
- Schumann, F.; Müller, A.; Koksch, M.; Müller, G.; Sewald, N. Are β-amino acids γ-turn mimetics? Exploring a new design principle for bioactive cyclopeptides. J. Am. Chem. Soc. 2000, 122, 12009–12010. [Google Scholar] [CrossRef]
- Urman, S.; Gaus, K.; Yang, Y.; Strijowski, U.; Sewald, N.; De Pol, S.; Reiser, O. The constrained amino acid β-Acc confers potency and selectivity to integrin ligands. Angew. Chem. Int. Ed. 2007, 46, 3976–3978. [Google Scholar] [CrossRef]
- Beumer, R.; Bubert, C.; Cabrele, C.; Vielhauer, O.; Pietzsch, M.; Reiser, O. The synthesis of diastereo- and enantiomerically pure β-aminocyclopropanecarboxylic acids. J. Org. Chem. 2000, 65, 8960–8969. [Google Scholar] [CrossRef]
- Allen, S.E.; Dokholyan, N.V.; Bowers, A.A. Dynamic Docking of Conformationally Constrained Macrocycles: Methods and Applications. ACS Chem. Biol. 2016, 11, 10–24. [Google Scholar] [CrossRef]
- Vasile, F.; Civera, M.; Belvisi, L.; Potenza, D.; Tiana, G. Thermodynamically-Weighted Conformational Ensemble of Cyclic RGD Peptidomimetics from NOE Data. J. Phys. Chem. B 2016, 120, 7098–7107. [Google Scholar] [CrossRef]
- Ohki, H.; Inamoto, Y.; Kawabata, K.; Kamimura, T.; Sakane, K. Synthesis and antifungal activity of FR109615 analogs. J. Antibiot. 1991, 44, 546–549. [Google Scholar] [CrossRef] [Green Version]
- Thorbeck, P.; Hjeds, H.; Schaumburg, K. Syntheses and 1H NMR spectroscopic investigations of some pyrrolidine carboxylic acids designed as potential glial GABA uptake inhibitors. Acta Chem. Scand. 1981, B35, 473–479. [Google Scholar] [CrossRef]
- Bunnage, M.E.; Davies, S.G.; Roberts, P.M.; Smith, A.D.; Withey, J.M. Asymmetric synthesis of the cis- and trans-stereoisomers of 4-aminopyrrolidine-3-carboxylic acid and 4-aminotetrahydrofuran-3-carboxylic acid. Org. Biomol. Chem. 2004, 2, 2763–2776. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Espinosa, J.F.; Gellman, S.H. 12-Helix formation in aqueous solution with short β-peptides containing pyrrolidine-based residues. J. Am. Chem. Soc. 2000, 122, 4821–4822. [Google Scholar] [CrossRef]
- Schmitt, M.A.; Choi, S.H.; Guzei, I.A.; Gellmann, S.H. New helical foldamers: Heterogeneous backbones with 1:2 and 2:1 α:β-amino acid residue patterns. J. Am. Chem. Soc. 2006, 128, 4538–4539. [Google Scholar] [CrossRef] [PubMed]
- Berlicki, Ł.; Pilsl, L.; Wéber, E.; Mándity, I.; Cabrele, C.; Martinek, T.A.; Fülöp, F.; Reiser, O. Unique α,β- and α,α,β,β-peptide foldamers based on cis-β-aminocyclopentanecarboxylic acid. Angew. Chem. Int. Ed. 2012, 51, 2208–2212. [Google Scholar] [CrossRef] [PubMed]
- Berlicki, Ł.; Kaske, M.; Gutiérrez-Abad, R.; Bernhardt, G.; Illa, O.; Ortuňo, R.M.; Cabrele, C.; Buschauer, A.; Reiser, O. Replacement of Thr32 and Gln34 in the C -terminal neuropeptide y fragment 25-36 by cis -cyclobutane and cis -cyclopentane β-amino acids shifts selectivity toward the Y4 receptor. J. Med. Chem. 2013, 56, 8422–8431. [Google Scholar] [CrossRef]
- Malešević, M.; Strijowski, U.; Bächle, D.; Sewald, N. An improved method for the solution cyclization of peptides under pseudo-high dilution conditions. J. Biotechnol. 2004, 112, 73–77. [Google Scholar] [CrossRef]
- Civera, M.; Arosio, D.; Bonato, F.; Manzoni, L.; Pignataro, L.; Zanella, S.; Gennari, C.; Piarulli, U.; Belvisi, L. Investigating the interaction of cyclic RGD peptidomimetics with αVβ6 integrin by biochemical and molecular docking studies. Cancers 2017, 9, 128.1. [Google Scholar] [CrossRef] [Green Version]
- Guzzetti, I.; Civera, M.; Vasile, F.; Arosio, D.; Tringali, C.; Piarulli, U.; Gennari, C.; Pignataro, L.; Belvisi, L.; Potenza, D. Insights into the Binding of Cyclic RGD Peptidomimetics to α5β1 Integrin by using Live-Cell NMR And Computational Studies. ChemistryOpen 2017, 6, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Borbély, A.; Figueras, E.; Martins, A.; Bodero, L.; Raposo Moreira Dias, A.; López Rivas, P.; Pina, A.; Arosio, D.; Gallinari, P.; Frese, M.; et al. Conjugates of Cryptophycin and RGD or isoDGR Peptidomimetics for Targeted Drug Delivery. ChemistryOpen 2019, 8, 737–742. [Google Scholar] [CrossRef] [Green Version]
- IMarelli, U.K.; Frank, A.O.; Wahl, B.; La Pietra, V.; Novellino, E.; Marinelli, L.; Herdtweck, E.; Groll, M.; Kessler, H. Receptor-Bound Conformation of Cilengitide Better Represented by Its Solution-State Structure than the Solid-State Structure. Chem. Eur. J. 2014, 20, 14201–14206. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Guida, W.C.; Still, W.C. An Internal Coordinate Monte Carlo Method for Searching Conformational Space. J. Am. Chem. Soc. 1989, 111, 4379–4386. [Google Scholar] [CrossRef]
- Still, W.C.; Tempczyk, A.; Hawley, R.C.; Hendrickson, T. Semianalytical Treatment of Solvation for Molecular Mechanics and Dynamics. J. Am. Chem. Soc. 1990, 112, 6127–6129. [Google Scholar] [CrossRef]
- Weinstock, D.S.; Narayanan, C.; Felts, A.K.; Andrec, M.; Levy, R.M.; Wu, K.P.; Baum, J. Distinguishing Among Structural Ensembles of the Gb1 Peptide: Remd Simulations and NMR experiments. J Am Chem Soc. 2007, 129, 4858–4859. [Google Scholar] [CrossRef] [Green Version]
- Davies, S.G.; Ichihara, O.; Walters, A.S. An expeditious asymmetric synthesis of (-)-(1R,2S)-cispentacin. Synlett 1993, 1993, 461–462. [Google Scholar] [CrossRef]
- Davies, S.G.; Ichihara, O.; Lenoir, I.; Walters, A.S. Asymmetric synthesis of (-)-(1R,2S)-cispentacin and related cis- and trans-2-amino cyclopentane- and cyclohexane-1-carboxylic acids. J. Chem. Soc. Perkin Trans. 1994, 1, 1411–1415. [Google Scholar] [CrossRef]
- Maestro Version 10.5; Schrödinger: New York, NY, USA, 2016.
- Macromodel, Version 11.1; Schrödinger: New York, NY, USA, 2016.
- Ponder, J.W.; Richards, F.M. An efficient newton-like method for molecular mechanics energy minimization of large molecules. J. Comput. Chem. 1987, 8, 1016–1024. [Google Scholar] [CrossRef]
- Xiong, J.P.; Stehle, T.; Zhang, R.; Joachimiak, A.; Frech, M.; Goodman, S.L.; Arnaout, M.A. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science 2002, 296, 151–155. [Google Scholar] [CrossRef]
- Glide, Version 7.0; Schrödinger: New York, NY, USA, 2016.







| Compound | αvβ3 IC50 [nM] [a] | α5β1 IC50 [nM] [a] |
|---|---|---|
| Cilengitide (1a) | 0.71 ± 0.06 | 14.4 ± 3.1 |
| cyclo-[-Arg-Gly-Asp-d-Phe-Val-] (1b) | 3.2 ± 1.3 | 166 ± 28 |
| cyclo-[-DKP-Arg-Gly-Asp-] (4) | 4.5 ± 1.1 | 532 ± 35 |
| cyclo-[-Arg-Gly-Asp-(1S,2R)-β-ACPC-Val-] (8) | 44.3 ± 4.0 | 3227 ± 1468 |
| cyclo-[-Arg-Gly-Asp-(1R,2S)-β-ACPC-Val-] (9) | 39.0 ± 1.1 | 468 ± 114 |
| cyclo-(-DKP-isoAsp-Gly-Arg-) (7) | 9.2 ± 1.1 | 1066 ± 228 |
| cyclo-[-isoAsp-Gly-Arg-(1R,2S)-β-ACPC-Val-] (10) | 5362 ± 281 | 2331 ± 134 |
| Compound | IC50 [μM] [a] |
|---|---|
| cyclo-(-Arg-Gly-Asp-d-Phe-Val-) (1b) | 5.1 ± 1.8 |
| cyclo-[-Arg-Gly-Asp-(1S,2R)-β-ACPC-Val-] (8) | 75 ± 32.5 |
| cyclo-[-Arg-Gly-Asp-(1R,2S)-β-ACPC-Val-] (9) | 124.5 ± 10.6 |
| cyclo-[-isoAsp-Gly-Arg-(1R,2S)-β-ACPC-Val-] (10) | >300 |
| Compound | NH-Gly δ (Δδ/ΔT) [a] | NH-Arg δ (Δδ/ΔT) [a] | NH-Asp δ (Δδ/ΔT) [a] | NH-ACPC δ (Δδ/ΔT) [a] | NH-Val δ (Δδ/ΔT) [a] |
|---|---|---|---|---|---|
| 8 | 8.37 (−7) | 9.10 (−9.5) | 7.73 (−2) | 7.33 (−4) | 7.92 (−6) |
| 9 | 8.62 (−7) | 8.30 (−5) | 8.38 (−8) | 7.59 (−5) | 7.52 (−6) |
| 10 | 8.04 (−3) | 8.44 (−7) | 8.42 (−7) | 7.91 (−4) | 7.74 (−9) |
| Compound | NOE Contact [a] | NOE Contact [a] |
|---|---|---|
| 8 | NH-Asp—NH-ACPC (s) | NH-Gly—NH-Asp (m) |
| 9 | NH-Asp—NH-ACPC (s) | |
| 10 | NH-Arg—NH-ACPC (s) | NH-Gly—NH-isoAsp (m) |
Sample Availability: Samples of the compounds are available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panzeri, S.; Arosio, D.; Gazzola, S.; Belvisi, L.; Civera, M.; Potenza, D.; Vasile, F.; Kemker, I.; Ertl, T.; Sewald, N.; et al. Cyclic RGD and isoDGR Integrin Ligands Containing cis-2-amino-1-cyclopentanecarboxylic (cis-β-ACPC) Scaffolds. Molecules 2020, 25, 5966. https://doi.org/10.3390/molecules25245966
Panzeri S, Arosio D, Gazzola S, Belvisi L, Civera M, Potenza D, Vasile F, Kemker I, Ertl T, Sewald N, et al. Cyclic RGD and isoDGR Integrin Ligands Containing cis-2-amino-1-cyclopentanecarboxylic (cis-β-ACPC) Scaffolds. Molecules. 2020; 25(24):5966. https://doi.org/10.3390/molecules25245966
Chicago/Turabian StylePanzeri, Silvia, Daniela Arosio, Silvia Gazzola, Laura Belvisi, Monica Civera, Donatella Potenza, Francesca Vasile, Isabell Kemker, Thomas Ertl, Norbert Sewald, and et al. 2020. "Cyclic RGD and isoDGR Integrin Ligands Containing cis-2-amino-1-cyclopentanecarboxylic (cis-β-ACPC) Scaffolds" Molecules 25, no. 24: 5966. https://doi.org/10.3390/molecules25245966