Engineering the Enantioselectivity of Yeast Old Yellow Enzyme OYE2y in Asymmetric Reduction of (E/Z)-Citral to (R)-Citronellal
Abstract
:1. Introduction
2. Results
2.1. OYE2y-Mediated Reduction of (E/Z)-Citral
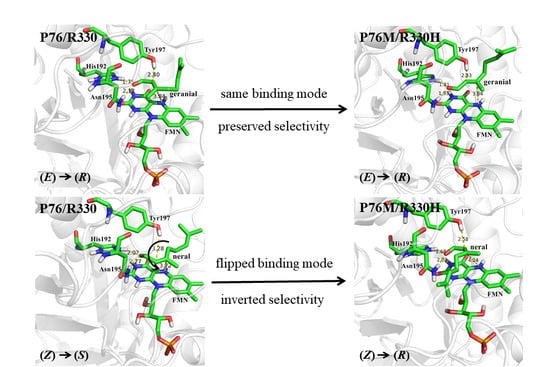
2.2. Identification of Key Residues for the Enantioselectivity of OYE2y
2.3. Site-Saturation Mutagenesis of R330 in OYE2y
2.4. Site-Saturation Mutagenesis of P76 in OYE2y
2.5. Evaluation of Double Substitution at Sites P76 and R330 of OYE2y
3. Discussion
4. Materials and Methods
4.1. Organisms and Chemicals
4.2. Preparation of (Z)-Citral and (E)-Citral
4.3. Cloning, Expression, and Purification of OYE2y
4.4. Construction of OYE2y Variants by Site-Directed Mutagenesis
4.5. Homology Modeling and Molecular Docking
4.6. Asymmetric Reduction of Citral Mediated by OYE2y or Its Variants
4.7. Analyses of GC and GC–MS
4.8. Nucleotide Sequence Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Itoh, H.; Maeda, H.; Yamada, S.; Hori, Y.; Mino, T.; Sakamoto, M. Kinetic resolution of citronellal by chiral aluminum catalysts: L-menthol synthesis from citral. Org. Chem. Front. 2014, 1, 1107–1115. [Google Scholar] [CrossRef]
- Lenardão, E.J.; Botteselle, G.V.; de Azambuja, F.; Perin, G.; Jacob, R.G. Citronellal as key compound in organic synthesis. Tetrahedron 2007, 63, 6671–6712. [Google Scholar] [CrossRef]
- Nie, Y.; Chuah, G.-K.; Jaenicke, S. Domino-cyclisation and hydrogenation of citronellal to menthol over bifunctional Ni/Zr-Beta and Zr-beta/Ni-MCM-41 catalysts. Chem. Commun. 2006, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Coffen, D.L.; Cohen, N.; Pico, A.M.; Schmid, R.; Sebastian, M.J.; Wang, F. A microbial lipase based stereoselective synthesis of (d)-α-tocopherol from (R)-citronellal and (S)-(6-hydroxy-2,5,7,8-tetramethylchroman-2-yl)acetic acid. Heterocycles 1994, 39, 527–552. [Google Scholar] [CrossRef]
- Eggersdorfer, M.; Laudert, D.; Létinois, U.; McClymont, T.; Medlock, J.; Netscher, T.; Bonrath, W. One hundred years of vitamins—a success story of the natural sciences. Angew. Chem. Int. Ed. 2012, 51, 12960–12990. [Google Scholar] [CrossRef] [PubMed]
- Tani, K.; Yamagata, T.; Akutagawa, S.; Kumobayashi, H.; Taketomi, T.; Takaya, H.; Miyashita, A.; Noyori, R.; Otsuka, S. Highly enantioselective isomerization of prochiral allylamines catalyzed by chiral diphosphine Rhodium(I) complexes: Preparation of optically active enamines. J. Am. Chem. Soc. 1984, 106, 5208–5217. [Google Scholar] [CrossRef]
- Zheng, L.; Lin, J.; Zhang, B.; Kuang, Y.; Wei, D. Identification of a yeast old yellow enzyme for highly enantioselective reduction of citral isomers to (R)-citronellal. Bioresour. Bioprocess. 2018, 5, 9. [Google Scholar] [CrossRef]
- Maeda, H.; Yamada, S.; Itoh, H.; Hori, Y. A dual catalyst system provides the shortest pathway for L-menthol synthesis. Chem. Commun. 2012, 48, 1772–1774. [Google Scholar] [CrossRef] [PubMed]
- Bougioukou, D.J.; Walton, A.Z.; Stewart, J.D. Towards preparative-scale, biocatalytic alkene reductions. Chem. Comm. 2010, 46, 8558–8560. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Hauer, B.; Rosche, B. Enzymatic reduction of the α,β-unsaturated carbon bond in citral. J. Mol. Catal. B Enzym. 2006, 38, 126–130. [Google Scholar] [CrossRef]
- Toogood, H.S.; Gardiner, J.M.; Scrutton, N.S. Biocatalytic reductions and chemical versatility of the old yellow enzyme family of flavoprotein oxidoreductases. ChemCatChem 2010, 2, 892–914. [Google Scholar] [CrossRef]
- Toogood, H.S.; Scrutton, N.S. New developments in ‘ene’-reductase catalysed biological hydrogenations. Curr. Opin. Chem. Biol. 2014, 19, 107–115. [Google Scholar] [CrossRef]
- Toogood, H.S.; Scrutton, N.S. Discovery, characterization, engineering, and applications of ene-reductases for industrial biocatalysis. ACS Catal. 2018, 8, 3532–3549. [Google Scholar] [CrossRef]
- Müller, A.; Hauer, B.; Rosche, B. Asymmetric alkene reduction by yeast old yellow enzymes and by a novel Zymomonas mobilis reductase. Biotechnol. Bioeng. 2007, 98, 22–29. [Google Scholar] [CrossRef]
- Brenna, E.; Gatti, F.G.; Monti, D.; Parmeggiani, F.; Serra, S. Stereochemical outcome of the biocatalysed reduction of activated tetrasubstituted olefins by old yellow enzymes 1–3. Adv. Synth. Catal. 2012, 354, 105–112. [Google Scholar] [CrossRef]
- Amato, E.D.; Stewart, J.D. Applications of protein engineering to members of the old yellow enzyme family. Biotechnol. Adv. 2015, 33, 624–631. [Google Scholar] [CrossRef]
- Kataoka, M.; Miyakawa, T.; Shimizu, S.; Tanokura, M. Enzymes useful for chiral compound synthesis: Structural biology, directed evolution, and protein engineering for industrial use. Appl. Microbiol. Biotechnol. 2016, 100, 5747–5757. [Google Scholar] [CrossRef]
- Li, R.; Wijma, H.J.; Song, L.; Cui, Y.; Otzen, M.; Tian, Y.; Du, J.; Li, T.; Niu, D.; Chen, Y.; et al. Computational redesign of enzymes for regio- and enantioselective hydroamination. Nature Chem. Biol. 2018, 14, 664–670. [Google Scholar] [CrossRef]
- Ying, X.; Zhang, J.; Wang, C.; Huang, M.; Ji, Y.; Cheng, F.; Yu, M.; Wang, Z.; Ying, M. Characterization of a carbonyl reductase from Rhodococcus erythropolis WZ010 and its variant Y54F for asymmetric synthesis of (S)-N-Boc-3-hydroxypiperidine. Molecules 2018, 23, 3117. [Google Scholar] [CrossRef]
- Daugherty, A.B.; Govindarajan, S.; Lutz, S. Improved biocatalysts from a synthetic circular permutation library of the flavin-dependent oxidoreductase old yellow enzyme. J. Am. Chem. Soc. 2013, 135, 14425–14432. [Google Scholar] [CrossRef]
- Quertinmont, L.T.; Lutz, S. Cell-free protein engineering of Old Yellow Enzyme 1 from Saccharomyces pastorianus. Tetrahedron 2016, 72, 7282–7287. [Google Scholar] [CrossRef]
- Padhi, S.K.; Bougioukou, D.J.; Stewart, J.D. Site-saturation mutagenesis of tryptophan 116 of Saccharomyces pastorianus old yellow enzyme uncovers stereocomplementary variants. J. Am. Chem. Soc. 2009, 131, 3271–3280. [Google Scholar] [CrossRef]
- Brenna, E.; Crotti, M.; Gatti, F.G.; Monti, D.; Parmeggiani, F.; Powell, R.W., III; Santangelo, S.; Stewart, J.D. Opposite enantioselectivity in the bioreduction of (Z)-β-aryl-bcyanoacrylates mediated by the tryptophan 116 mutants of old yellow enzyme 1: Synthetic approach to (R)- and (S)-β-aryl-glactams. Adv. Synth. Catal. 2015, 357, 1849–1860. [Google Scholar] [CrossRef]
- Pompeu, Y.A.; Sullivan, B.; Stewart, J.D. X-ray crystallography reveals how subtle changes control the orientation of substrate binding in an alkene reductase. ACS Catal. 2013, 3, 2376–2390. [Google Scholar] [CrossRef]
- Walton, A.Z.; Sullivan, B.; Patterson-Orazem, A.C.; Stewart, J.D. Residues controlling facial selectivity in an alkene reductase and semirational alterations to create stereocomplementary variants. ACS Catal. 2015, 4, 2307–2318. [Google Scholar] [CrossRef]
- Rüthlein, E.; Classen, T.; Dobnikar, L.; Schölzel, M.; Pietruszka, J. Finding the selectivity switch—A rational approach towards stereocomplementary variants of the ene reductase YqjM. Adv. Synth. Catal. 2015, 357, 1775–1786. [Google Scholar] [CrossRef]
- Yin, B.; Deng, J.; Lim, L.; Yuan, Y.A.; Wei, D. Structural insights into stereospecific reduction of α, β-unsaturated carbonyl substrates by old yellow enzyme from Gluconobacter oxydans. Biosci. Biotechnol. Biochem. 2015, 79, 410–421. [Google Scholar] [CrossRef]
- Kress, N.; Rapp, J.; Hauer, B. Enantioselective reduction of citral isomers in NCR ene reductase: Analysis of an active site mutant library. ChemBioChem 2017, 18, 717–720. [Google Scholar] [CrossRef]
- Hall, M.; Stueckler, C.; Hauer, B.; Stuermer, R.; Friedrich, T.; Breuer, M.; Kroutil, W.; Faber, K. Asymmetric bioreduction of activated C=C bonds using Zymomonas mobilis NCR enoate reductase and old yellow enzymes OYE 1-3 from yeasts. Eur. J. Org. Chem. 2008, 1511–1516. [Google Scholar] [CrossRef]
- Wolken, W.A.M.; ten Have, R.; van der Werf, M.J. Amino acid-catalyzed conversion of citral: Cis-trans isomerization and its conversion into 6-methyl-5-hepten-2-one and acetaldehyde. J. Agric. Food Chem. 2000, 48, 5401–5405. [Google Scholar] [CrossRef]
- Tsuboi, S.; Ishii, N.; Sakai, T.; Tari, I.; Utaka, M. Oxidation of alcohols with electrolytic manganese dioxide. Its application fro the synthesis of insect pheromones. Bull. Chem. Soc. Jpn. 1990, 63, 1888–1893. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Hall, M.; Hauer, B.; Stuermer, R.; Kroutil, W.; Faber, K. Asymmetric whole-cell bioreduction of an α,β-unsaturated aldehyde (citral): Competing prim-alcohol dehydrogenase and C-C lyase activities. Tetrahedron: Asymmetry 2006, 17, 3058–3062. [Google Scholar] [CrossRef]
- Cheng, F.; Tang, X.; Kardashliev, T. Transcription factor-based biosensors in high-throughput screening: Advances and applications. Biotechnol. J. 2018, 13, 1700648. [Google Scholar] [CrossRef]
- Cheng, F.; Zhu, L.; Schwaneberg, U. Directed evolution 2.0: Improving and deciphering enzyme properties. Chem. Commun. 2015, 51, 9760–9772. [Google Scholar] [CrossRef]
- Deng, J.; Yao, Z.; Chen, K.; Yuan, Y.A.; Lin, J.; Wei, D. Towards the computational design and engineering of enzyme enantioselectivity: A case study by a carbonyl reductase from Gluconobacter oxydans. J. Biotechnol. 2016, 217, 31–40. [Google Scholar] [CrossRef]
- Sullivan, B.; Walton, A.Z.; Stewart, J.D. Library construction and evaluation for site saturation mutagenesis. Enzyme Microb. Technol. 2013, 53, 70–77. [Google Scholar] [CrossRef]
- Valetti, F.; Gilardi, G. Improvement of biocatalysts for industrial and environmental purposes by saturation mutagenesis. Biomolecules 2013, 3, 778–811. [Google Scholar] [CrossRef]
- Yu, M.; Huang, M.; Song, Q.; Shao, J.; Ying, X. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase from Rhodococcus erythropolis WZ010. Molecules 2015, 20, 7156–7173. [Google Scholar] [CrossRef]
- Fox, K.M.; Karplus, P.A. Old yellow enzyme at 2 Å resolution: Overall structure, ligand binding, and comparison with related flavoproteins. Structure 1994, 2, 1089–1105. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An antomated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef]
- Breithaupt, C.; Strassner, J.; Breitinger, U.; Huber, R.; Macheroux, P.; Schaller, A.; Clausen, T. X-Ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE. Structure 2001, 9, 419–429. [Google Scholar] [CrossRef]
- Fraaije, M.W.; Mattevi, A. Flavoenzymes: Diverse catalysts with recurrent features. Trends Biochem. Sci. 2000, 25, 126–132. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors |






| Enzyme | (E)-Citral | (Z)-Citral | (E/Z)-Citral | |||
|---|---|---|---|---|---|---|
| e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | |
| S13G | 83.19 ± 1.57 (R) | 95.12 ± 2.10 | 45.29 ± 1.46 (S) | 94.60 ± 1.22 | 37.35 ± 1.63 (R) | 96.34 ± 1.87 |
| S59A | 80.69 ± 2.21 (R) | 90.32 ± 1.54 | 30.96 ± 1.25 (S) | 90.88 ± 2.23 | 41.66 ± 1.59 (R) | 93.48 ± 1.07 |
| V289I | 75.18 ± 1.70 (R) | 94.88 ± 1.81 | 37.45 ± 1.55 (S) | 91.71 ± 2.10 | 35.52 ± 2.42 (R) | 90.78 ± 0.93 |
| R330H | 88.08 ± 1.39 (R) | 71.23 ± 0.85 | 71.92 ± 1.34 (R) | 64.12 ± 1.33 | 86.88 ± 1.36 (R) | 52.83 ± 0.75 |
| P76M | 86.22 ± 0.75 (R) | 80.39 ± 0.64 | 4.76 ± 1.50 (S) | 55.19 ± 0.95 | 57.60 ± 0.92 (R) | 61.53 ± 0.89 |
| OYE2y | 82.87 ± 0.98 (R) | 92.20 ± 1.07 | 32.66 ± 1.77 (S) | 88.65 ± 1.49 | 38.13 ± 1.55 (R) | 89.51 ± 1.68 |
| Enzyme | (E)-Citral | (Z)-Citral | (E/Z)-Citral | |||
|---|---|---|---|---|---|---|
| e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | |
| R330H | 88.08 ± 1.39 (R) | 71.23 ± 0.85 | 71.92 ± 1.34 (R) | 64.12 ± 1.33 | 86.88 ± 1.36 (R) | 52.83 ± 0.75 |
| R330D | 92.42 ± 2.30 (R) | 63.12 ± 1.93 | 73.56 ± 0.51 (R) | 36.41 ± 1.94 | 80.30 ± 1.85 (R) | 62.70 ± 0.85 |
| R330W | 95.05 ± 1.85 (R) | 47.32 ± 1.11 | 72.19 ± 1.09 (R) | 53.38 ± 1.12 | 79.71 ± 1.03 (R) | 70.98 ± 1.15 |
| R330I | 87.38 ± 1.11 (R) | 31.45 ± 0.40 | 20.93 ± 1.21 (R) | 13.19 ± 0.23 | 74.52 ± 0.69 (R) | 28.17 ± 0.68 |
| R330L | 89.64 ± 0.79 (R) | 75.14 ± 1.25 | 45.59 ± 0.75 (R) | 55.72 ± 0.59 | 72.58 ± 1.32 (R) | 67.45 ± 1.21 |
| R330F | 89.40 ± 1.05 (R) | 89.86 ± 1.74 | 57.68 ± 0.86 (R) | 78.39 ± 1.34 | 72.50 ± 1.46 (R) | 80.49 ± 1.69 |
| R330E | 89.22 ± 0.61 (R) | 80.92 ± 0.80 | 47.80 ± 1.14 (R) | 47.82 ± 1.11 | 69.42 ± 2.34 (R) | 72.46 ± 1.02 |
| R330A | 88.10 ± 1.37 (R) | 83.10 ± 2.11 | 51.07 ± 1.23 (R) | 49.12 ± 1.63 | 69.42 ± 1.11 (R) | 69.61 ± 0.72 |
| R330T | 85.74 ± 2.33 (R) | 83.33 ± 2.06 | 27.97 ± 0.48 (R) | 43.83 ± 0.87 | 69.12 ± 2.32 (R) | 46.61 ± 0.29 |
| R330N | 92.15 ± 2.49 (R) | 90.55 ± 1.85 | 41.87 ± 0.73 (R) | 72.44 ± 1.61 | 68.19 ± 1.56 (R) | 78.89 ± 1.98 |
| R330V | 89.66 ± 1.50 (R) | 91.10 ± 1.01 | 24.39 ± 1.55 (R) | 65.15 ± 1.69 | 67.68 ± 1.42 (R) | 75.77 ± 1.35 |
| R330S | 89.60 ± 1.48 (R) | 85.62 ± 1.38 | 33.95 ± 0.67 (R) | 69.67 ± 1.92 | 64.39 ± 2.05 (R) | 71.80 ± 1.57 |
| R330C | 87.52 ± 1.56 (R) | 80.50 ± 1.47 | 33.80 ± 1.41 (R) | 63.79 ± 1.27 | 64.35 ± 2.33 (R) | 70.16 ± 0.86 |
| R330K | 86.18 ± 0.84 (R) | 84.82 ± 2.19 | 5.45 ± 0.59 (R) | 63.69 ± 1.51 | 61.53 ± 1.76 (R) | 80.64 ± 0.43 |
| R330Q | 84.81 ± 1.41 (R) | 90.74 ± 2.55 | 10.90 ± 0.84 (R) | 76.88 ± 0.66 | 61.17 ± 0.71 (R) | 85.23 ± 1.44 |
| R330G | 92.39 ± 1.66 (R) | 10.96 ± 0.29 | 5.07 ± 1.22 (R) | 8.49 ± 0.15 | 60.97 ± 1.56 (R) | 14.82 ± 0.33 |
| R330M | 87.88 ± 1.23 (R) | 92.40 ± 1.77 | 0.37 ± 0.65 (R) | 85.18 ± 2.08 | 58.40 ± 1.08 (R) | 86.71 ± 1.37 |
| R330Y | 87.03 ± 1.12 (R) | 96.61 ± 2.16 | 40.92 ± 1.53 (S) | 88.24 ± 1.89 | 38.65 ± 1.11 (R) | 91.56 ± 1.72 |
| R330P b | / | / | / | / | / | / |
| OYE2y | 82.87 ± 0.98 (R) | 92.20 ± 1.07 | 32.66 ± 1.77 (S) | 88.65 ± 1.49 | 38.13 ± 1.55 (R) | 89.51 ± 1.68 |
| Enzyme | (E)-Citral | (Z)-Citral | (E/Z)-Citral | |||
|---|---|---|---|---|---|---|
| e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | |
| P76C | 85.39 ± 2.70 (R) | 65.32 ± 0.76 | 37.50 ± 1.55 (R) | 38.54 ± 1.61 | 69.92 ± 2.46 (R) | 49.65 ± 1.40 |
| P76S | 81.23 ± 1.79 (R) | 81.84 ± 0.62 | 10.19 ± 0.73 (S) | 48.15 ± 0.88 | 62.80 ± 0.97 (R) | 64.48 ± 0.90 |
| P76M | 86.22 ± 0.75 (R) | 80.39 ± 0.64 | 4.76 ± 1.50 (S) | 55.19 ± 0.95 | 57.60 ± 1.92 (R) | 61.53 ± 0.89 |
| P76G | 85.52 ± 2.55 (R) | 83.94 ± 0.78 | 3.63 ± 1.47 (S) | 57.54 ± 1.84 | 53.19 ± 0.81 (R) | 78.04 ± 1.52 |
| P76N | 86.45 ± 1.33 (R) | 85.36 ± 0.49 | 13.05 ± 0.67 (S) | 69.39 ± 1.43 | 49.59 ± 1.55 (R) | 72.59 ± 0.77 |
| P76A | 86.55 ± 0.91 (R) | 90.57 ± 1.98 | 21.21 ± 1.95 (S) | 88.96 ± 0.87 | 41.88 ± 0.95 (R) | 90.43 ± 2.22 |
| P76V | 87.54 ± 1.55 (R) | 92.50 ± 2.51 | 30.15 ± 1.67 (S) | 82.30 ± 2.07 | 40.49 ± 1.01 (R) | 87.37 ± 1.28 |
| P76T | 86.12 ± 1.35 (R) | 93.79 ± 2.26 | 28.10 ± 1.69 (S) | 91.24 ± 2.12 | 37.05 ± 1.13 (R) | 91.50 ± 0.94 |
| P76L | 84.28 ± 1.62 (R) | 90.95 ± 1.83 | 38.01 ± 2.27 (S) | 88.36 ± 1.46 | 35.05 ± 1.64 (R) | 88.46 ± 1.32 |
| P76I | 86.10 ± 1.03 (R) | 94.76 ± 1.74 | 40.23 ± 1.81 (S) | 89.95 ± 1.33 | 34.51 ± 1.17 (R) | 91.88 ± 2.45 |
| P76Y b | / | / | / | / | / | / |
| P76Q b | / | / | / | / | / | / |
| P76D b | / | / | / | / | / | / |
| P76E b | / | / | / | / | / | / |
| P76R b | / | / | / | / | / | / |
| P76H b | / | / | / | / | / | / |
| P76F b | / | / | / | / | / | / |
| P76W b | / | / | / | / | / | / |
| P76K b | / | / | / | / | / | / |
| OYE2y | 82.87 ± 0.98 (R) | 92.20 ± 1.07 | 32.66 ± 1.77 (S) | 88.65 ± 1.49 | 38.13 ± 1.55 (R) | 89.51 ± 1.68 |
| Enzyme | (E)-Citral | (Z)-Citral | (E/Z)-Citral | |||
|---|---|---|---|---|---|---|
| e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | e.e. (%) | Yield (%) | |
| P76C | 85.39 ± 2.70 (R) | 65.32 ± 0.76 | 37.50 ± 1.55 (R) | 38.54 ± 1.61 | 69.92 ± 2.46 (R) | 49.65 ± 1.40 |
| P76C/R330H | 88.00 ± 2.04 (R) | 78.09 ± 1.16 | 76.16 ± 0.88 (R) | 54.53 ± 0.45 | 82.32 ± 1.22 (R) | 57.74 ± 0.84 |
| P76C/R330D | 88.77 ± 1.81 (R) | 80.00 ± 1.52 | 76.45 ± 1.03 (R) | 52.23 ± 1.25 | 81.70 ± 2.18 (R) | 63.28 ± 1.36 |
| P76C/R330W | 89.53 ± 2.23 (R) | 70.30 ± 1.87 | 70.32 ± 1.16 (R) | 56.49 ± 0.68 | 81.44 ± 1.31 (R) | 65.31 ± 1.72 |
| P76C/R330C | 87.43 ± 1.37 (R) | 90.10 ± 1.96 | 49.17 ± 2.44 (R) | 51.14 ± 0.79 | 77.37 ± 1.04 (R) | 66.19 ± 1.42 |
| R330H | 88.08 ± 1.39 (R) | 71.23 ± 0.85 | 71.92 ± 1.34 (R) | 64.12 ± 1.33 | 86.88 ± 1.36 (R) | 52.83 ± 0.75 |
| P76M/R330H | >99 (R) | 73.88 ± 2.66 | 75.01 ± 1.59 (R) | 13.99 ± 0.23 | >99 (R) | 15.83 ± 0.45 |
| P76G/R330H | >99 (R) | 64.09 ± 1.32 | 77.60 ± 2.06 (R) | 12.64 ± 0.31 | >99 (R) | 11.61 ± 0.38 |
| P76S/R330H | >99 (R) | 64.74 ± 1.57 | 74.74 ± 1.84 (R) | 8.90 ± 0.16 | >99 (R) | 9.12 ± 0.22 |
| OYE2y | 82.87 ± 0.98 (R) | 92.20 ± 1.07 | 32.66 ± 1.77 (S) | 88.65 ± 1.49 | 38.13 ± 1.55 (R) | 89.51 ± 1.68 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, X.; Yu, S.; Huang, M.; Wei, R.; Meng, S.; Cheng, F.; Yu, M.; Ying, M.; Zhao, M.; Wang, Z. Engineering the Enantioselectivity of Yeast Old Yellow Enzyme OYE2y in Asymmetric Reduction of (E/Z)-Citral to (R)-Citronellal. Molecules 2019, 24, 1057. https://doi.org/10.3390/molecules24061057
Ying X, Yu S, Huang M, Wei R, Meng S, Cheng F, Yu M, Ying M, Zhao M, Wang Z. Engineering the Enantioselectivity of Yeast Old Yellow Enzyme OYE2y in Asymmetric Reduction of (E/Z)-Citral to (R)-Citronellal. Molecules. 2019; 24(6):1057. https://doi.org/10.3390/molecules24061057
Chicago/Turabian StyleYing, Xiangxian, Shihua Yu, Meijuan Huang, Ran Wei, Shumin Meng, Feng Cheng, Meilan Yu, Meirong Ying, Man Zhao, and Zhao Wang. 2019. "Engineering the Enantioselectivity of Yeast Old Yellow Enzyme OYE2y in Asymmetric Reduction of (E/Z)-Citral to (R)-Citronellal" Molecules 24, no. 6: 1057. https://doi.org/10.3390/molecules24061057