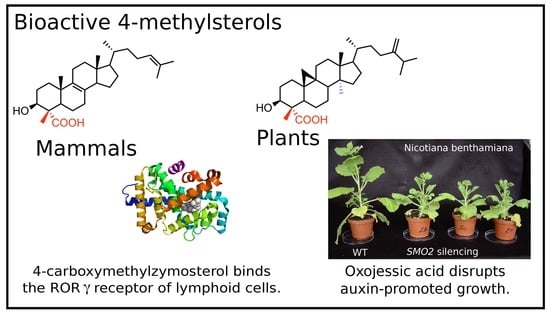
Metabolism and Biological Activities of 4-Methyl-Sterols
Abstract
:1. An Introduction to 4-Methylsterols
2. Some Crucial Milestones in Deciphering the Sterol-Demethylation Process and Functions of C4-SBIs in Mammals
3. Saccharomyces cerevisiae, a Versatile Model for Sterol Genetics and Auxotrophy Studies
4. The Plant-Specific Sterol-C4-Demethylation Process and Its Influence upon Development
5. Caenorhabditis elegans: A Sterol Auxotroph with an Extraordinary C4-Methylation Capacity
6. Bacteria Evolved Their Specific C4-Demethylation Enzymes
7. Inhibitors of C4-SBIs Accumulation In Vivo, Canonical and Non-Canonical C4-SBIs, and Conjugated forms
8. Concluding Remarks
Supplementary Materials
Funding
Conflicts of Interest
References
- Itoh, T.; Tamura, T.; Matsumoto, T. Methylsterol compositions of 19 vegetable oils. J. Am. Oil Chem. Soc. 1973, 50, 300–303. [Google Scholar] [CrossRef]
- Phillips, K.M.; Ruggio, D.M.; Toivo, J.I.; Swank, M.A.; Simpkins, A.H. Free and Esterified Sterol Composition of Edible Oils and Fats. J. Food Compost. Anal. 2002, 15, 123–142. [Google Scholar] [CrossRef]
- Villette, C.; Berna, A.; Compagnon, V.; Schaller, H. Plant Sterol Diversity in Pollen from Angiosperms. Lipids 2015, 50, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Moreau, R.A.; Nystrom, L.; Whitaker, B.D.; Winkler-Moser, J.K.; Baer, D.J.; Gebauer, S.K.; Hicks, K.B. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and health-promoting uses. Prog. Lipid Res. 2018, 70, 35–61. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.A.; Venkatramesh, M.; Nes, W.D. Developmental regulation of sterol biosynthesis in Zea mays. Lipids 1995, 30, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Benveniste, P. Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 2004, 55, 429–457. [Google Scholar] [CrossRef]
- Nes, W.D. Biosynthesis of cholesterol and other sterols. Chem. Rev. 2011, 111, 6423–6451. [Google Scholar] [CrossRef]
- Schaller, H. Sterol and steroid biosynthesis and metabolism in plants and microorganisms. In Comprehensive Natural Products II: Chemistry and Biology; Mander, L.N., Hung-Wen, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2010; Volume 1, pp. 755–787. [Google Scholar]
- Abe, I.; Rohmer, M.; Prestwich, G.D. Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem. Rev. 1993, 93, 2189–2206. [Google Scholar] [CrossRef]
- Gaylor, J.L.; Moir, N.J.; Seifried, H.E.; Jefcoate, C.R. Assay and isolation of a cyanide-binding protein of rat liver microsomes. J. Biol. Chem. 1970, 245, 5511–5513. [Google Scholar]
- Trzaskos, J.M.; Bowen, W.D.; Fisher, G.J.; Billheimer, J.T.; Gaylor, J.L. Microsomal enzymes of cholesterol biosynthesis from lanosterol: A progress report. Lipids 1982, 17, 250–256. [Google Scholar] [CrossRef]
- Trzaskos, J.M.; Bowen, W.D.; Shafiee, A.; Fischer, R.T.; Gaylor, J.L. Cytochrome P450-dependent oxidation of lanosterol in cholesterol biosynthesis. Microsomal transport and C32-demethylation. J. Biol. Chem. 1984, 259, 13402–13412. [Google Scholar] [PubMed]
- Aoyama, Y.; Okikawa, T.; Yoshida, Y. Evidence for the presence of cytochrome P-450 functional in lanosterol 14α-demethylation in microsomes of aerobically grown respiring yeast. Biochim. Biophys. Acta 1981, 665, 596–601. [Google Scholar] [CrossRef]
- Rahier, A.; Taton, M. The 14α-demethylation of obtusifoliol by a cytochrome P-450 monooxygenase from higher plants microsomes. Biochem. Biophys. Res. Commun. 1986, 140, 1064–1072. [Google Scholar] [CrossRef]
- Rahier, A.; Taton, M. Plant Sterol Biosynthesis Inhibitors—The 14-Demethylation Steps, Their Enzymology and Inhibition. Biochem. Soc. Trans. 1990, 18, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Schaller, H.; Goh, C.H.; Kwon, M.; Choe, S.; An, C.S.; Durst, F.; Feldmann, K.A.; Feyereisen, R. Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 2005, 138, 2033–2047. [Google Scholar] [CrossRef]
- Schrick, K.; Mayer, U.; Horrichs, A.; Kuhnt, C.; Bellini, C.; Dangl, J.; Schmidt, J.; Jurgens, G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000, 14, 1471–1484. [Google Scholar]
- Lorenz, R.T.; Parks, L.W. Cloning, sequencing, and disruption of the gene encoding sterol C-14 reductase in Saccharomyces cerevisiae. DNA Cell Biol. 1992, 11, 685–692. [Google Scholar] [CrossRef]
- Roberti, R.; Bennati, A.M.; Galli, G.; Caruso, D.; Maras, B.; Aisa, C.; Beccari, T.; Della Fazia, M.A.; Servillo, G. Cloning and expression of sterol Δ14-reductase from bovine liver. Eur. J. Biochem. 2002, 269, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Waterham, H.R.; Koster, J.; Romeijn, G.J.; Hennekam, R.C.; Vreken, P.; Andersson, H.C.; FitzPatrick, D.R.; Kelley, R.I.; Wanders, R.J. Mutations in the 3β-hydroxysterol Δ24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am. J. Hum. Genet. 2001, 69, 685–694. [Google Scholar] [CrossRef]
- Pascal, S.; Taton, M.; Rahier, A. Plant sterol biosynthesis. Identification and characterization of two distinct microsomal oxidative enzymatic systems involved in sterol C4-demethylation. J. Biol. Chem. 1993, 268, 11639–11654. [Google Scholar]
- Bouvier, F.; Rahier, A.; Camara, B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 2005, 44, 357–429. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Bard, M. Erg28p is a key protein in the yeast sterol biosynthetic enzyme complex. J. Lipid Res. 2005, 46, 1991–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darnet, S. Biosynthèse des stérols: Biochimie et analyse moléculaire des stérol-4α-méthyl-oxydases de plantes; Université de Strasbourg: Strasbourg, France, 2004. [Google Scholar]
- Santori, F.R.; Huang, P.; van de Pavert, S.A.; Douglass, E.F., Jr.; Leaver, D.J.; Haubrich, B.A.; Keber, R.; Lorbek, G.; Konijn, T.; Rosales, B.N.; et al. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. 2015, 21, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Volkman, J.K.; Rijpstra, W.I.C.; de Leeuw, J.W.; Mansour, M.P.; Jackson, A.E.; Blackburn, S.I. Sterols of four dinoflagellates from the genus Prorocentrum. Phytochemistry 1999, 52, 659–668. [Google Scholar] [CrossRef]
- Bouvier, P.; Rohmer, M.; Benveniste, P.; Ourisson, G. Δ8(14)-steroids in the bacterium Methylococcus capsulatus. Biochem. J. 1976, 159, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Volkman, J.K. Sterols and other triterpenoids: Source specificity and evolution of biosynthetic pathways. Org. Geochem. 2005, 36, 139–159. [Google Scholar] [CrossRef]
- Fowler, M.G.; Douglas, A.G. Saturated hydrocarbon biomarkers in oils of Late Precambrian age from Eastern Siberia. Org. Geochem. 1987, 11, 201–213. [Google Scholar] [CrossRef]
- Ourisson, G. Pecularities of Sterol Biosynthesis in Plants. J. Plant Physiol. 1994, 143, 434–439. [Google Scholar] [CrossRef]
- Bloch, K.E. Sterol structure and membrane function. CRC Crit. Rev. Biochem. 1983, 14, 47–92. [Google Scholar] [CrossRef]
- Gas-Pascual, E.; Berna, A.; Bach, T.J.; Schaller, H. Plant oxidosqualene metabolism: Cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana. PLoS ONE 2014, 9, e109156. [Google Scholar] [CrossRef]
- Barton, D.H.R. Triterpenoids. Part III. cycloartenone, a triterpenoid ketone. J. Chem. Soc. 1951, 1444. [Google Scholar] [CrossRef]
- Goad, L.J. Aspects of Phytosterol Biosynthesis. In Terpenoids in Plants; Pridham, J.B., Ed.; Academic Press: London, UK, 1967; p. 159. [Google Scholar]
- Lenton, J.R.; John Goad, L.; Goodwin, T.W. Sitosterol biosynthesis in Hordeum vulgare. Phytochemistry 1975, 14, 1523–1528. [Google Scholar] [CrossRef]
- Dahl, C.E.; Dahl, J.S.; Bloch, K. Effect of alkyl-substituted precursors of cholesterol on artificial and natural membranes and on the viability of Mycoplasma capricolum. Biochemistry 1980, 19, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Volkman, J.K. Sterols in microorganisms. Appl. Microbiol. Biotechnol. 2003, 60, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, H.; Ohnishi, M.; Ogawa, S. Distribution of Unusual Cholesterol Precursors, 4-Methyl- and 4, 4-Dimethylsterols with Δ8 Unsaturation, in Gonads of Marine Archaeogastropods. J. Oleo Sci. 2013, 62, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.; Jackson-Lewis, V.; Wong, L.C.; Shui, G.H.; Goh, A.X.; Kesavapany, S.; Jenner, A.M.; Fivaz, M.; Przedborski, S.; Wenk, M.R. Lanosterol induces mitochondrial uncoupling and protects dopaminergic neurons from cell death in a model for Parkinson’s disease. Cell Death Differ. 2012, 19, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Hubler, Z.; Allimuthu, D.; Bederman, I.; Elitt, M.S.; Madhavan, M.; Allan, K.C.; Shick, H.E.; Garrison, E.; Karl, M.T.; Factor, D.C.; et al. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature 2018, 560, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Rozman, D.; Cotman, M.; Frangež, R. Lanosterol 14α-demethylase and MAS sterols in mammalian gametogenesis. Mol. Cell. Endocrinol. 2002, 187, 179–187. [Google Scholar] [CrossRef]
- Byskov, A.G.; Andersen, C.Y.; Leonardsen, L. Role of meiosis activating sterols, MAS, in induced oocyte maturation. Mol. Cell. Endocrinol. 2002, 187, 189–196. [Google Scholar] [CrossRef]
- Byskov, A.G.; Andersen, C.Y.; Nordholm, L.; Thogersen, H.; Xia, G.; Wassmann, O.; Andersen, J.V.; Guddal, E.; Roed, T. Chemical structure of sterols that activate oocyte meiosis. Nature 1995, 374, 559–562. [Google Scholar] [CrossRef]
- Grondahl, C.; Hansen, T.H.; Marky-Nielsen, K.; Ottesen, J.L.; Hyttel, P. Human oocyte maturation in vitro is stimulated by meiosis-activating sterol. Hum. Reprod. 2000, 15 (Suppl. 5), 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsche, M.A.; McDonald, J.G.; Hobbs, H.H.; Cohen, J.C. Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. Elife 2015, 4, e07999. [Google Scholar] [CrossRef] [PubMed]
- Alonso, F.; Cirigliano, A.M.; Davola, M.E.; Cabrera, G.M.; Garcia Linares, G.E.; Labriola, C.; Barquero, A.A.; Ramirez, J.A. Multicomponent synthesis of 4,4-dimethyl sterol analogues and their effect on eukaryotic cells. Steroids 2014, 84, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Blume, T.; Guttzeit, M.; Kuhnke, J.; Zorn, L. Two syntheses of FF-MAS. Org. Lett. 2003, 5, 1837–1839. [Google Scholar] [CrossRef] [PubMed]
- Konig, A.; Happle, R.; Bornholdt, D.; Engel, H.; Grzeschik, K.H. Mutations in the NSDHL gene, encoding a 3β-hydroxysteroid dehydrogenase, cause CHILD syndrome. Am. J. Med. Genet. 2000, 90, 339–346. [Google Scholar] [CrossRef]
- McLarren, K.W.; Severson, T.M.; du Souich, C.; Stockton, D.W.; Kratz, L.E.; Cunningham, D.; Hendson, G.; Morin, R.D.; Wu, D.; Paul, J.E.; et al. Hypomorphic temperature-sensitive alleles of NSDHL cause CK syndrome. Am. J. Hum. Genet. 2010, 87, 905–914. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Kratz, L.E.; Michel, J.J.; Vallejo, A.N.; Ferris, L.; Kelley, R.I.; Hoover, J.J.; Jukic, D.; Gibson, K.M.; Wolfe, L.A.; et al. Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J. Clin. Investig. 2011, 121, 976–984. [Google Scholar] [CrossRef] [Green Version]
- Hannich, J.T.; Entchev, E.V.; Mende, F.; Boytchev, H.; Martin, R.; Zagoriy, V.; Theumer, G.; Riezman, I.; Riezman, H.; Knolker, H.J.; et al. Methylation of the sterol nucleus by STRM-1 regulates dauer larva formation in Caenorhabditis elegans. Dev. Cell 2009, 16, 833–843. [Google Scholar] [CrossRef]
- Mialoundama, A.S.; Jadid, N.; Brunel, J.; Di Pascoli, T.; Heintz, D.; Erhardt, M.; Mutterer, J.; Bergdoll, M.; Ayoub, D.; Van Dorsselaer, A.; et al. Arabidopsis ERG28 tethers the sterol C4-demethylation complex to prevent accumulation of a biosynthetic intermediate that interferes with polar auxin transport. Plant Cell 2013, 25, 4879–4893. [Google Scholar] [CrossRef]
- Fisher, R.T.; Trzaskos, J.M.; Magolda, R.L.; Ko, S.S.; Brosz, C.S.; Larsen, B. Lanosterol 14α-Methyl Demethylase. J. Biol. Chem. 1991, 10, 6124–6132. [Google Scholar]
- Stromstedt, M.; Rozman, D.; Waterman, M.R. The ubiquitously expressed human CYP51 encodes lanosterol 14α-demethylase, a cytochrome P450 whose expression is regulated by oxysterols. Arch. Biochem. Biophys. 1996, 329, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Bennati, A.M.; Castelli, M.; Della Fazia, M.A.; Beccari, T.; Caruso, D.; Servillo, G.; Roberti, R. Sterol dependent regulation of human TM7SF2 gene expression: Role of the encoded 3β-hydroxysterol Δ14-reductase in human cholesterol biosynthesis. Biochim. Biophys. Acta 2006, 1761, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Bennati, A.M.; Schiavoni, G.; Franken, S.; Piobbico, D.; Della Fazia, M.A.; Caruso, D.; De Fabiani, E.; Benedetti, L.; Cusella De Angelis, M.G.; Gieselmann, V.; et al. Disruption of the gene encoding 3β-hydroxysterol Δ14-reductase (Tm7sf2) in mice does not impair cholesterol biosynthesis. FEBS J. 2008, 275, 5034–5047. [Google Scholar] [CrossRef]
- Gatticchi, L.; Cerra, B.; Scarpelli, P.; Macchioni, L.; Sebastiani, B.; Gioiello, A.; Roberti, R. Selected cholesterol biosynthesis inhibitors produce accumulation of the intermediate FF-MAS that targets nucleus and activates LXRalpha in HepG2 cells. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.L.; Zhao, C.; Turner, E.; Schlieker, C. The Lamin B receptor is essential for cholesterol synthesis and perturbed by disease-causing mutations. Elife 2016, 5, e16011. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.A., Jr.; Lindberg, M.; Bloch, K. On the demethylation of lanosterol to cholesterol. J. Biol. Chem. 1957, 226, 941–956. [Google Scholar] [PubMed]
- Lindberg, M.; Gautschi, F.; Bloch, K.E. Ketonic Intermediates in the demethylation of lanosterol. J. Biol. Chem. 1957, 238, 1661–1664. [Google Scholar]
- Pudles, J.; Bloch, K. Conversion of 4-hydroxymethylene-∆7-cholesten-3-one to cholesterol. J. Biol. Chem. 1960, 235, 3417–3420. [Google Scholar]
- Swindell, A.C.; Gaylor, J.L. Investigation of the component reactions of oxidative sterol demethylation. Formation and metabolism of 3-ketosteroid intermediates. J. Biol. Chem. 1968, 243, 5546–5555. [Google Scholar]
- Miller, W.L.; Gaylor, J.L. Investigation of the component reactions of oxidative sterol demethylation. Oxidation of a 4,4-dimethyl sterol to a 4 beta-methyl-4 alpha-carboxylic acid during cholesterol biosynthesis. J. Biol. Chem. 1970, 245, 5375–5381. [Google Scholar]
- Miller, W.L.; Brady, D.R.; Gaylor, J.L. Investigation of the component reactions of oxidative demethylation of sterols: Metabolism of 4alpha-hydroxymethyl steroids. J. Biol. Chem. 1971, 246, 5147–5153. [Google Scholar] [PubMed]
- Rahimtula, A.D.; Gaylor, J.L. Investigation of the component reactions of oxidative sterol demethylation. Partial purification of a microsomal sterol 4α-carboxylic acid decarboxylase. J. Biol. Chem. 1972, 247, 9–15. [Google Scholar] [PubMed]
- Billheimer, J.T.; Alcorn, M.; Gaylor, J.L. Solubilization and partial purification of a microsomal 3-ketosteroid reductase of cholesterol biosynthesis. Arch. Biochem. Biophys. 1981, 211, 430–438. [Google Scholar] [CrossRef]
- Nelson, J.A.; Kahn, S.; Spencer, T.A.; Sharpless, K.B.; Clayton, R.B. Some Aspects of Substrate-Specificity in Biological Demethylation at C4 of Steroids. Bioorg. Chem. 1975, 4, 363–376. [Google Scholar] [CrossRef]
- Gaylor, J.L.; Miyake, Y.; Yamano, T. Stoichiometry of 4-methyl sterol oxidase of rat liver microsomes. J. Biol. Chem. 1975, 250, 7159–7167. [Google Scholar] [PubMed]
- Marijanovic, Z.; Laubner, D.; Moller, G.; Gege, C.; Husen, B.; Adamski, J.; Breitling, R. Closing the gap: Identification of human 3-ketosteroid reductase, the last unknown enzyme of mammalian cholesterol biosynthesis. Mol. Endocrinol. 2003, 17, 1715–1725. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Dangel, A.W.; Kelley, R.I.; Zhao, W.; Denny, P.; Botcherby, M.; Cattanach, B.; Peters, J.; Hunsicker, P.R.; Mallon, A.M.; et al. The gene mutated in bare patches and striated mice encodes a novel 3β-hydroxysteroid dehydrogenase. Nat. Genet. 1999, 22, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Ottolenghi, C.; Daizadeh, I.; Ju, A.; Kossida, S.; Renault, G.; Jacquet, M.; Fellous, A.; Gilbert, W.; Veitia, R. The genomic structure of c14orf1 is conserved across eukarya. Mamm. Genome 2000, 11, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, R.A. DRUG DISCOVERY. A new dawn for cataracts. Science 2015, 350, 636–637. [Google Scholar] [CrossRef] [PubMed]
- Makley, L.N.; McMenimen, K.A.; DeVree, B.T.; Goldman, J.W.; McGlasson, B.N.; Rajagopal, P.; Dunyak, B.M.; McQuade, T.J.; Thompson, A.D.; Sunahara, R.; et al. Pharmacological chaperone for alpha-crystallin partially restores transparency in cataract models. Science 2015, 350, 674–677. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, X.J.; Zhu, J.; Xi, Y.B.; Yang, X.; Hu, L.D.; Ouyang, H.; Patel, S.H.; Jin, X.; Lin, D.; et al. Lanosterol reverses protein aggregation in cataracts. Nature 2015, 523, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, P.; Barigali, A.; Kadaskar, J.; Borgohain, S.; Mishra, D.C.; Ramanjulu, R.; Minija, C.K. Effect of lanosterol on human cataract nucleus. Indian J. Ophthalmol. 2015, 63, 888–890. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Hu, L.D.; Yao, K.; Yan, Y.B. Lanosterol and 25-hydroxycholesterol dissociate crystallin aggregates isolated from cataractous human lens via different mechanisms. Biochem. Biophys. Res. Commun. 2018, 506, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhu, M.; Kang, L.; Tu, Y.; Li, L.; Zhang, R.; Qin, B.; Yang, M.; Guan, H. Lanosterol Synthase Pathway Alleviates Lens Opacity in Age-Related Cortical Cataract. J. Ophthalmol. 2018, 2018, 4125893. [Google Scholar] [CrossRef] [PubMed]
- Araldi, E.; Fernandez-Fuertes, M.; Canfran-Duque, A.; Tang, W.; Cline, G.W.; Madrigal-Matute, J.; Pober, J.S.; Lasuncion, M.A.; Wu, D.; Fernandez-Hernando, C.; et al. Lanosterol Modulates TLR4-Mediated Innate Immune Responses in Macrophages. Cell Rep. 2017, 19, 2743–2755. [Google Scholar] [CrossRef] [PubMed]
- Song, B.L.; Javitt, N.B.; DeBose-Boyd, R.A. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005, 1, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.D.; McDonald, J.G.; Bruick, R.K.; DeBose-Boyd, R.A. Hypoxia stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase through accumulation of lanosterol and hypoxia-inducible factor-mediated induction of insigs. J. Biol. Chem. 2007, 282, 27436–27446. [Google Scholar] [CrossRef]
- Schiavoni, G.; Bennati, A.M.; Castelli, M.; Della Fazia, M.A.; Beccari, T.; Servillo, G.; Roberti, R. Activation of TM7SF2 promoter by SREBP-2 depends on a new sterol regulatory element, a GC-box, and an inverted CCAAT-box. Biochim. Biophys. Acta 2010, 1801, 587–592. [Google Scholar] [CrossRef]
- Subramanian, G.; Chaudhury, P.; Malu, K.; Fowler, S.; Manmode, R.; Gotur, D.; Zwerger, M.; Ryan, D.; Roberti, R.; Gaines, P. Lamin B receptor regulates the growth and maturation of myeloid progenitors via its sterol reductase domain: Implications for cholesterol biosynthesis in regulating myelopoiesis. J. Immunol. 2012, 188, 85–102. [Google Scholar] [CrossRef]
- Bellezza, I.; Roberti, R.; Gatticchi, L.; Del Sordo, R.; Rambotti, M.G.; Marchetti, M.C.; Sidoni, A.; Minelli, A. A novel role for Tm7sf2 gene in regulating TNFalpha expression. PLoS ONE 2013, 8, e68017. [Google Scholar] [CrossRef]
- Bellezza, I.; Gatticchi, L.; del Sordo, R.; Peirce, M.J.; Sidoni, A.; Roberti, R.; Minelli, A. The loss of Tm7sf gene accelerates skin papilloma formation in mice. Sci. Rep. 2015, 5, 9471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartoli, D.; Piobbico, D.; Bellet, M.M.; Bennati, A.M.; Roberti, R.; Della Fazia, M.A.; Servillo, G. Impaired cell proliferation in regenerating liver of 3β-hydroxysterol Δ14-reductase (TM7SF2) knock-out mice. Cell Cycle 2016, 15, 2164–2173. [Google Scholar] [CrossRef] [PubMed]
- Offiah, A.C.; Mansour, S.; Jeffrey, I.; Nash, R.; Whittock, N.; Pyper, R.; Bewley, S.; Clayton, P.T.; Hall, C.M. Greenberg dysplasia (HEM) and lethal X linked dominant Conradi-Hünermann chondrodysplasia punctata (CDPX2): Presentation of two cases with overlapping phenotype. J. Med. Genet. 2003, 40, e129. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, C.R.; Rimoin, D.L.; Gruber, H.E.; DeSa, D.J.; Reed, M.; Lachman, R.S. A new autosomal recessive lethal chondrodystrophy with congenital hydrops. Am. J. Med. Genet. 1988, 29, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, E.; Sirchia, F.; Bosco, M.; Sobreira, N.L.M.; Baylor-Hopkins Center for Mendelian Genomics; Grosso, E.; Brussino, A.; Brusco, A. A novel case of Greenberg dysplasia and genotype-phenotype correlation analysis for LBR pathogenic variants: An instructive example of one gene-multiple phenotypes. Am. J. Med. Genet. A 2018. [Google Scholar] [CrossRef] [PubMed]
- Gaudy-Marqueste, C.; Roll, P.; Esteves-Vieira, V.; Weiller, P.J.; Grob, J.J.; Cau, P.; Levy, N.; De Sandre-Giovannoli, A. LBR mutation and nuclear envelope defects in a patient affected with Reynolds syndrome. J. Med. Genet. 2010, 47, 361–370. [Google Scholar] [CrossRef]
- Sukhanova, A.; Gorin, A.; Serebriiskii, I.G.; Gabitova, L.; Zheng, H.; Restifo, D.; Egleston, B.L.; Cunningham, D.; Bagnyukova, T.; Liu, H.; et al. Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov. 2013, 3, 96–111. [Google Scholar] [CrossRef]
- He, M.; Smith, L.D.; Chang, R.; Li, X.; Vockley, J. The role of sterol-C4-methyl oxidase in epidermal biology. Biochim. Biophys. Acta 2014, 1841, 331–335. [Google Scholar] [CrossRef] [Green Version]
- Mi, X.B.; Luo, M.X.; Guo, L.L.; Zhang, T.D.; Qiu, X.W. CHILD Syndrome: Case Report of a Chinese Patient and Literature Review of the NAD[P]H Steroid Dehydrogenase-Like Protein Gene Mutation. Pediatr. Dermatol. 2015, 32, e277–e282. [Google Scholar] [CrossRef] [PubMed]
- Seeger, M.A.; Paller, A.S. The role of abnormalities in the distal pathway of cholesterol synthesis in the Congenital Hemidysplasia with Ichthyosiform erythroderma and Limb Defects (CHILD) syndrome. Biochim. Biophys. Acta 2014, 1841, 345–352. [Google Scholar] [CrossRef]
- Stottmann, R.W.; Turbe-Doan, A.; Tran, P.; Kratz, L.E.; Moran, J.L.; Kelley, R.I.; Beier, D.R. Cholesterol metabolism is required for intracellular hedgehog signal transduction in vivo. PLoS Genet. 2011, 7, e1002224. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.; DeBarber, A.E.; Bir, N.; Binkley, L.; Merkens, L.S.; Steiner, R.D.; Herman, G.E. Analysis of hedgehog signaling in cerebellar granule cell precursors in a conditional Nsdhl allele demonstrates an essential role for cholesterol in postnatal CNS development. Hum. Mol. Genet. 2015, 24, 2808–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, R. Sterol metabolism and SREBP activation. Arch. Biochem. Biophys. 2010, 501, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Moss, G.P. Nomenclature of steroids (Recommendations 1989). Pure Appl. Chem. 1989, 61, 1783–1822. [Google Scholar] [CrossRef]
- Daum, G.; Lees, N.D.; Bard, M.; Dickson, R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 1998, 14, 1471–1510. [Google Scholar] [CrossRef]
- Lees, N.D.; Bard, M.; Kirsch, D.R. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 33–47. [Google Scholar]
- Li, L.; Kaplan, J. Characterization of yeast methyl sterol oxidase (ERG25) and identification of a human homologue. J. Biol. Chem. 1996, 271, 16927–16933. [Google Scholar] [CrossRef] [PubMed]
- Bard, M.; Bruner, D.A.; Pierson, C.A.; Lees, N.D.; Biermann, B.; Frye, L.; Koegel, C.; Barbuch, R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc. Natl. Acad. Sci. USA 1996, 93, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Ishizuka, H.; Beppu, T. Cloning, nucleotide sequence, and transcriptional analysis of the NAD(P)-dependent cholesterol dehydrogenase gene from a Nocardia sp. and its hyperexpression in Streptomyces spp. Appl. Environ. Microbiol. 1991, 57, 1386–1393. [Google Scholar] [PubMed]
- Gachotte, D.; Barbuch, R.; Gaylor, J.; Nickel, E.; Bard, M. Characterization of the Saccharomyces cerevisiae ERG26 gene encoding the C-3 sterol dehydrogenase (C-4 decarboxylase) involved in sterol biosynthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 13794–13799. [Google Scholar] [CrossRef]
- Aaron, K.E.; Pierson, C.A.; Lees, N.D.; Bard, M. The Candida albicans ERG26 gene encoding the C-3 sterol dehydrogenase (C-4 decarboxylase) is essential for growth. FEMS Yeast Res. 2001, 1, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Gachotte, D.; Sen, S.E.; Eckstein, J.; Barbuch, R.; Krieger, M.; Ray, B.D.; Bard, M. Characterization of the Saccharomyces cerevisiae ERG27 gene encoding the 3-keto reductase involved in C-4 sterol demethylation. Proc. Natl. Acad. Sci. USA 1999, 96, 12655–12660. [Google Scholar] [CrossRef] [PubMed]
- Gachotte, D.; Eckstein, J.; Barbuch, R.; Hughes, T.; Roberts, C.; Bard, M. A novel gene conserved from yeast to humans is involved in sterol biosynthesis. J. Lipid Res. 2001, 42, 150–154. [Google Scholar] [PubMed]
- Baudry, K.; Swain, E.; Rahier, A.; Germann, M.; Batta, A.; Rondet, S.; Mandala, S.; Henry, K.; Tint, G.S.; Edlind, T.; et al. The effect of the erg26-1 mutation on the regulation of lipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 12702–12711. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Valachovic, M.; Randall, S.K.; Nickels, J.T.; Bard, M. Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 9739–9744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, D.M.; Chen, O.S.; Li, L.; Kaplan, J.; Bhuiyan, S.A.; Natarajan, S.K.; Bard, M.; Cox, J.E. Altered sterol metabolism in budding yeast affects mitochondrial iron-sulfur (Fe-S) cluster synthesis. J. Biol. Chem. 2018, 293, 10782–10795. [Google Scholar] [CrossRef] [PubMed]
- Gachotte, D.; Pierson, C.A.; Lees, N.D.; Barbuch, R.; Koegel, C.; Bard, M. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc. Natl. Acad. Sci. USA 1997, 94, 11173–11178. [Google Scholar] [CrossRef] [Green Version]
- Wachtler, V.; Rajagopalan, S.; Balasubramanian, M.K. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 2003, 116, 867–874. [Google Scholar] [CrossRef]
- Germann, M.; Gallo, C.; Donahue, T.; Shirzadi, R.; Stukey, J.; Lang, S.; Ruckenstuhl, C.; Oliaro-Bosso, S.; McDonough, V.; Turnowsky, F.; et al. Characterizing sterol defect suppressors uncovers a novel transcriptional signaling pathway regulating zymosterol biosynthesis. J. Biol. Chem. 2005, 280, 35904–35913. [Google Scholar] [CrossRef]
- Swain, E.; Baudry, K.; Stukey, J.; McDonough, V.; Germann, M.; Nickels, J.T., Jr. Sterol-dependent regulation of sphingolipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 26177–26184. [Google Scholar] [CrossRef]
- Lv, J.M.; Hu, D.; Gao, H.; Kushiro, T.; Awakawa, T.; Chen, G.D.; Wang, C.X.; Abe, I.; Yao, X.S. Biosynthesis of helvolic acid and identification of an unusual C-4-demethylation process distinct from sterol biosynthesis. Nat. Commun. 2017, 8, 1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, A.L.; Lee, C.Y.; Bien, C.M.; Espenshade, P.J. 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J. Biol. Chem. 2007, 282, 24388–24396. [Google Scholar] [CrossRef] [PubMed]
- Rahier, A.; Smith, M.; Taton, M. The role of cytochrome b5 in 4α-methyl-oxidation and C5(6) desaturation of plant sterol precursors. Biochem. Biophys. Res. Commun. 1997, 236, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Darnet, S.; Rahier, A. Plant sterol biosynthesis: Identification of two distinct families of sterol 4α-methyl oxidases. Biochem. J. 2004, 378, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Darnet, S.; Bard, M.; Rahier, A. Functional identification of sterol-4α-methyl oxidase cDNAs from Arabidopsis thaliana by complementation of a yeast erg25 mutant lacking sterol-4α-methyl oxidation. FEBS Lett. 2001, 508, 39–43. [Google Scholar] [CrossRef]
- Sonawane, P.D.; Pollier, J.; Panda, S.; Szymanski, J.; Massalha, H.; Yona, M.; Unger, T.; Malitsky, S.; Arendt, P.; Pauwels, L.; et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 2016, 3, 16205. [Google Scholar] [CrossRef]
- Rahier, A.; Darnet, S.; Bouvier, F.; Camara, B.; Bard, M. Molecular and enzymatic characterizations of novel bifunctional 3β-hydroxysteroid dehydrogenases/C-4 decarboxylases from Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 27264–27277. [Google Scholar] [CrossRef]
- Lung, S.C.; Liao, P.; Yeung, E.C.; Hsiao, A.S.; Xue, Y.; Chye, M.L. Arabidopsis ACYL-COA-BINDING PROTEIN1 interacts with STEROL C4-METHYL OXIDASE1-2 to modulate gene expression of homeodomain-leucine zipper IV transcription factors. New Phytol. 2018, 218, 183–200. [Google Scholar] [CrossRef]
- Lung, S.C.; Liao, P.; Yeung, E.C.; Hsiao, A.S.; Xue, Y.; Chye, M.L. Acyl-CoA-Binding Protein ACBP1 Modulates Sterol Synthesis during Embryogenesis. Plant Physiol. 2017, 174, 1420–1435. [Google Scholar] [CrossRef] [Green Version]
- Lange, I.; Poirier, B.C.; Herron, B.K.; Lange, B.M. Comprehensive Assessment of Transcriptional Regulation Facilitates Metabolic Engineering of Isoprenoid Accumulation in Arabidopsis. Plant Physiol. 2015, 169, 1595–1606. [Google Scholar] [CrossRef]
- Burger, C.; Rondet, S.; Benveniste, P.; Schaller, H. Virus-induced silencing of sterol biosynthetic genes: Identification of a Nicotiana tabacum L. obtusifoliol-14α-demethylase (CYP51) by genetic manipulation of the sterol biosynthetic pathway in Nicotiana benthamiana L. J. Exp. Bot. 2003, 54, 1675–1683. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, M.; Schmit, A.C.; Heintz, D.; Schaller, H.; Ohta, D. Diversification of sterol methyltransferase enzymes in plants and a role for beta-sitosterol in oriented cell plate formation and polarized growth. Plant J. 2015, 84, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Schaller, H.; Bouvier-Navé, P.; Benveniste, P. Overexpression of an Arabidopsis cDNA Encoding a Sterol-C241-Methyltransferase in Tobacco Modifies the Ratio of 24-Methyl Cholesterol to Sitosterol and Is Associated with Growth Reduction. Plant Physiol. 1998, 118, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, A.; Bronner, R.; Benveniste, P.; Schaller, H. The ratio of campesterol to sitosterol with modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2-1. Plant J. 2001, 25, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sasvari, Z.; Nagy, P.D. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J. Virol. 2010, 84, 2270–2281. [Google Scholar] [CrossRef]
- Kim, B.; Kim, G.; Fujioka, S.; Takatsuto, S.; Choe, S. Overexpression of 3beta-hydroxysteroid dehydrogenases/C-4 decarboxylases causes growth defects possibly due to abnormal auxin transport in Arabidopsis. Mol. Cells 2012, 34, 77–84. [Google Scholar] [CrossRef]
- Simon-Plas, F.; Perraki, A.; Bayer, E.; Gerbeau-Pissot, P.; Mongrand, S. An update on plant membrane rafts. Curr. Opin. Plant Biol. 2011, 14, 642–649. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, S.; Nie, X.; Boutte, Y.; Grison, M.; Li, P.; Kuang, S.; Men, S. Sterol Methyl Oxidases Affect Embryo Development via Auxin-Associated Mechanisms. Plant Physiol. 2016, 171, 468–482. [Google Scholar] [CrossRef]
- Klahre, U.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Yokota, T.; Nomura, T.; Yoshida, S.; Chua, N.H. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 1998, 10, 1677–1690. [Google Scholar] [CrossRef]
- Choe, S.; Dilkes, B.P.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Feldmann, K.A. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 1998, 10, 231. [Google Scholar]
- Choe, S.W.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Tissier, C.P.; Gregory, B.D.; Ross, A.S.; Tanaka, A.; Yoshida, S.; Tax, F.E.; et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 1999, 11, 207–221. [Google Scholar] [PubMed]
- Choe, S.; Dilkes, B.P.; Gregory, B.D.; Ross, A.S.; Yuan, H.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Tanaka, A.; Yoshida, S.; et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999, 119, 897–907. [Google Scholar] [CrossRef]
- Silvestro, D.; Andersen, T.G.; Schaller, H.; Jensen, P.E. Plant Sterol Metabolism. Δ7-Sterol-C5-Desaturase (STE1/DWARF7), Δ5,7-Sterol-Δ7-Reductase (DWARF5) and Δ24-Sterol-Δ24-Reductase (DIMINUTO/DWARF1) Show Multiple Subcellular Localizations in Arabidopsis thaliana (Heynh) L. PLoS ONE 2013, 8, e56429. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, V.; Friml, J.; Grebe, M.; van den Toorn, A.; Palme, K.; Scheres, B. Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 2003, 15, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Men, S.; Boutte, Y.; Ikeda, Y.; Li, X.; Palme, K.; Stierhof, Y.D.; Hartmann, M.A.; Moritz, T.; Grebe, M. Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 2008, 10, 237–244. [Google Scholar] [CrossRef]
- Butcher, R.A. Small-molecule pheromones and hormones controlling nematode development. Nat. Chem. Biol. 2017, 13, 577–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matyash, V.; Entchev, E.V.; Mende, F.; Wilsch-Brauninger, M.; Thiele, C.; Schmidt, A.W.; Knolker, H.J.; Ward, S.; Kurzchalia, T.V. Sterol-derived hormone(s) controls entry into diapause in Caenorhabditis elegans by consecutive activation of DAF-12 and DAF-16. PLoS Biol. 2004, 2, e280. [Google Scholar] [CrossRef]
- Lee, A.K.; Banta, A.B.; Wei, J.H.; Kiemle, D.J.; Feng, J.; Giner, J.L.; Welander, P.V. C-4 sterol demethylation enzymes distinguish bacterial and eukaryotic sterol synthesis. Proc. Natl. Acad. Sci. USA 2018, 115, 5884–5889. [Google Scholar] [CrossRef]
- Wollam, J.; Magomedova, L.; Magner, D.B.; Shen, Y.; Rottiers, V.; Motola, D.L.; Mangelsdorf, D.J.; Cummins, C.L.; Antebi, A. The Rieske oxygenase DAF-36 functions as a cholesterol 7-desaturase in steroidogenic pathways governing longevity. Aging Cell 2011, 10, 879–884. [Google Scholar] [CrossRef]
- Poklepovich, T.J.; Urtasun, N.; Miranda, M.V.; Nusblat, A.D.; Nudel, C.B. Expression and functional characterization of a C-7 cholesterol desaturase from Tetrahymena thermophila in an insect cell line. Steroids 2015, 96, 132–139. [Google Scholar] [CrossRef]
- Najle, S.R.; Nusblat, A.D.; Nudel, C.B.; Uttaro, A.D. The Sterol-C7 desaturase from the ciliate Tetrahymena thermophila is a Rieske Oxygenase, which is highly conserved in animals. Mol. Biol. Evol. 2013, 30, 1630–1643. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C.; Brown, A.J.P.; Gow, N.A.R. Antifungal agents: Mechanisms of action. Trends Microbiol. 2003, 11, 272–279. [Google Scholar] [CrossRef]
- Warrilow, A.G.; Parker, J.E.; Kelly, D.E.; Kelly, S.L. Azole affinity of sterol 14α-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob. Agents Chemother. 2013, 57, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Binder, U.; Bracher, F.; Giera, M. Antifungal drug testing by combining minimal inhibitory concentration testing with target identification by gas chromatography-mass spectrometry. Nat. Protoc. 2017, 12, 947–963. [Google Scholar] [CrossRef] [PubMed]
- Jachak, G.R.; Ramesh, R.; Sant, D.G.; Jorwekar, S.U.; Jadhav, M.R.; Tupe, S.G.; Deshpande, M.V.; Reddy, D.S. Silicon Incorporated Morpholine Antifungals: Design, Synthesis, and Biological Evaluation. ACS Med. Chem. Lett. 2015, 6, 1111–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuchta, T.; Bartkova, K.; Kubinec, R. Ergosterol depletion and 4-methyl sterols accumulation in the yeast Saccharomyces cerevisiae treated with an antifungal, 6-amino-2-n-pentylthiobenzothiazole. Biochem. Biophys. Res. Commun. 1992, 189, 85–91. [Google Scholar] [CrossRef]
- Kuchta, T.; Leka, C.; Farkas, P.; Bujdakova, H.; Belajova, E.; Russell, N.J. Inhibition of sterol 4-demethylation in Candida albicans by 6-amino-2-n-pentylthiobenzothiazole, a novel mechanism of action for an antifungal agent. Antimicrob. Agents Chemother. 1995, 39, 1538–1541. [Google Scholar] [CrossRef] [PubMed]
- Fabry, S.; Gaborova, S.; Bujdakova, H.; Klobusicky, M.; Vollekova, A.; Kuchta, T. Inhibition of germ tube formation, filamentation and ergosterol biosynthesis in Candida albicans treated with 6-amino-2-n-pentylthiobenzothiazole. Folia Microbiol. (Praha) 1999, 44, 523–526. [Google Scholar] [CrossRef]
- Darnet, S.; Rahier, A. Enzymological properties of sterol-C4-methyl-oxidase of yeast sterol biosynthesis. Biochim. Biophys. Acta 2003, 1633, 106–117. [Google Scholar] [CrossRef]
- Nose, H.; Fushimi, H.; Seki, A.; Sasaki, T.; Watabe, H.; Hoshiko, S. PF1163A, a Novel Antifungal Agent, Inhibit Ergosterol Biosynthesis at C-4 Sterol Methyl Oxidase. J. Antibiot. 2002, 55, 969–974. [Google Scholar] [CrossRef]
- Ekhato, I.V.; Robinson, C.H. Synthesis of New Nitro and Amino Sterols—Potential Inhibitors of 4-Methyl Sterol Oxidase. J. Chem. Soc.-Perkin Trans. 1 1988, 3239–3242. [Google Scholar] [CrossRef]
- Ekhato, I.V.; Robinson, C.H. Synthesis of Novel 4α-Substituted Sterols. J. Org. Chem. 1989, 54, 1327–1331. [Google Scholar] [CrossRef]
- Singh, D.K.; Porter, T.D. Inhibition of sterol 4α-methyl oxidase is the principal mechanism by which garlic decreases cholesterol synthesis. J. Nutr. 2006, 136, 759s–764s. [Google Scholar] [CrossRef]
- Lindenthal, B.; Holleran, A.L.; Aldaghlas, T.A.; Ruan, B.; Schroepfer, G.J., Jr.; Wilson, W.K.; Kelleher, J.K. Progestins block cholesterol synthesis to produce meiosis-activating sterols. FASEB J. 2001, 15, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, F.; Hayashi, H. Identification of intermediates after inhibition of cholesterol synthesis by aminotriazole treatment in vivo. Biochim. Biophys. Acta 1991, 1086, 115–124. [Google Scholar] [CrossRef]
- Hashimoto, F.; Hayashi, H. Peroxisomal cholesterol synthesis in vivo: Accumulation of 4-methyl intermediate sterols after aminotriazole inhibition of cholesterol synthesis. Biochim. Biophys. Acta 1994, 1214, 11–19. [Google Scholar] [CrossRef]
- Helliwell, S.B.; Karkare, S.; Bergdoll, M.; Rahier, A.; Leighton-Davis, J.R.; Fioretto, C.; Aust, T.; Filipuzzi, I.; Frederiksen, M.; Gounarides, J.; et al. FR171456 is a specific inhibitor of mammalian NSDHL and yeast Erg26p. Nat. Commun. 2015, 6, 8613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debieu, D.; Bach, J.; Hugon, M.; Malosse, C.; Leroux, P. The hydroxyanilide fenhexamid, a new sterol biosynthesis inhibitor fungicide efficient against the plant pathogenic fungus Botryotinia fuckeliana (Botrytis cinerea). Pest Manag. Sci. 2001, 57, 1060–1067. [Google Scholar] [CrossRef]
- Trzaskos, J.M.; Fischer, R.T.; Favata, M.F. Mechanistic studies of lanosterol C-32 demethylation. Conditions which promote oxysterol intermediate accumulation during the demethylation process. J. Biol. Chem. 1986, 261, 16937–16942. [Google Scholar] [PubMed]
- Ori, K.; Koroda, M.; Mimaki, Y.; Sakagami, H.; Sashida, Y. Lanosterol and tetranorlanosterol glycosides from the bulbs of Muscari paradoxum. Phytochemistry 2003, 64, 1351–1359. [Google Scholar] [CrossRef]
- Ragasa, C.Y.; Tiu, F.; Rideout, J.A. New cycloartenol esters from Ixora coccinea. Nat. Prod. Res. 2004, 18, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Gallo, C.; d’Ippolito, G.; Nuzzo, G.; Sardo, A.; Fontana, A. Autoinhibitory sterol sulfates mediate programmed cell death in a bloom-forming marine diatom. Nat. Commun. 2017, 8, 1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massé, R.; Huang, Y.S.; Eid, K.; Laliberté, C.; Davignon, J. Plasma methyl sterol sulfates in familial hypercholesterolemia after partial ileal bypass. Can. J. Biochem. 1982, 60, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, Y.; Hao, L.Y.; Liu, X.; Lesch, C.A.; Sanchez, B.M.; Wendling, J.M.; Morgan, R.W.; Aicher, T.D.; Carter, L.L.; et al. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORgamma agonists. Nat. Chem. Biol. 2015, 11, 141–147. [Google Scholar] [CrossRef] [PubMed]





| ID | Common Name | IUPAC |
|---|---|---|
| M1 | lanosterol | lanosta-8,24-dien-3β-ol |
| M2 | FF-MAS | 4,4-dimethyl-5α-cholesta-8,14,24-trien-3β-ol |
| M3 | T-MAS, 4,4-dimethylzymosterol | 4,4-dimethyl-5α-cholesta-8,24-dien-3β-ol |
| M4 | oxojessic acid, CMMC | 4α-carboxy-4β,14α-dimethyl-9β,19-cyclo-5α-ergosta-24(241)-en-3β-ol |
| M5 | 4α-formyl-lanosterol | 4α-formyl-4β,14α-methyl-cholesta-8,24-dien-3β-ol |
| M6 | zymosterone | 5α-cholesta-8,24-dien-3-one |
| M7 | dinosterol | 4α,23,24-trimethyl-5α-cholesta-22-en-3β-ol |
| M8 | 4α-methyl-24-ethylcholestane | 4α,24-methyl-cholestan-3β-ol |
| M9 | 4α-hydroxymethyl-4β-methyl-zymosterol | 4α-hydroxymethyl-4β-methyl-cholesta-8,24-dien-3β-ol |
| M10 | 4α-formyl-4β-methylzymosterol | 4α-formyl-4β-methyl-cholesta-8,24-dien-3β-ol |
| M11 | 4α-carboxy-4β-methylzymosterol | 4α-carboxy-4β-methyl-cholesta-8,24-dien-3β-ol |
| M12 | 3-keto-4α-methylzymosterol | 4α-methyl-5α-cholesta-8,24-dien-3-one |
| M13 | 4α-methylzymosterol | 4α-methyl-5α-cholesta-8,24-dien-3β-ol |
| M14 | 4α-hydroxymethylzymosterol | 4α-hydroxymethyl-5α-cholesta-8,24-dien-3β-ol |
| M15 | 4α-formylzymosterol | 4α-formyl-5α-cholesta-8,24-dien-3β-ol |
| M16 | 4α-carboxyzymosterol | 4α-carboxy-5α-cholesta-8,24-dien-3β-ol |
| M17 | zymosterol | 5α-cholesta-8,24-dien-3β-ol |
| M18 | cholesterol | cholest-5-en-3β-ol |
| M19 | ergosterol | ergosta-5,7,22E-trien-3β-ol |
| M20 | - | 4α-hydroxymethyl-4β,14α-methyl-cholesta-8,24-dien-3β-ol |
| M21 | - | 4α-carboxy-4β,14α-methyl-cholesta-8,24-dien-3β-ol |
| M22 | cycloartenol | 9β,19-cyclo-lanost-24-en-3β-ol |
| M23 | 24-methylenecycloartanol | 24-methylene-9β,19-cyclo-lanost-3β-ol |
| M24 | 4-hydroxymethyl-24-methylenecycloartanol | 4α-hydroxymethyl-24-methylene-9β,19-cyclo-lanost-3β-ol |
| M25 | 4-formyl-24-methylenecycloartanol | 4α-formyl-24-methylene-9β,19-cyclo-lanost-3β-ol |
| M26 | cycloeucalenone | 24-methylene-9β,19-cyclo-lanost-3-one |
| M27 | 24-methylenelophenol | 4α-methyl-24-methylene-cholest-7-en-3β-ol |
| M28 | 24-ethylidenelophenol | 4α-methyl-24Z-ethylidene-cholest-7-en-3β-ol |
| M29 | 4-hydroxymethyl-24-ethylidenelophenol | 4α-hydroxymethyl-24Z-ethylidene-cholest-7-en-3β-ol |
| M30 | 4-formyl-24-ethylidenelophenol | 4α-formyl-24Z-ethylidene-cholest-7-en-3β-ol |
| M31 | 4-carboxy-24-ethylidenelophenol | 4α-carboxy-24Z-ethylidene-cholest-7-en-3β-ol |
| M32 | avenasterone | 24Z-ethylidene-cholest-7-en-3-one |
| M33 | Δ7-avenasterol | 24Z-ethylidene-cholest-7-en-3β-ol |
| M34 | sitosterol | stigmast-5-en-3β-ol |
| M35 | 4-hydroxymethyl-24-methylenelophenol | 4α-hydroxy-24Z-methylene-cholest-7-en-3β-ol |
| M36 | 4-formyl-24-methylenelophenol | 4α-formyl-24Z-methylene-cholest-7-en-3β-ol |
| M37 | 4-carboxy-24-methylenelophenol | 4α-carboxy-24Z-methylene-cholest-7-en-3β-ol |
| M38 | episterone | 24-methylene-cholest-7-en-3β-one |
| M39 | episterol | 24Z-methylene-cholest-7-en-3β-ol |
| M40 | campesterol | campest-5-en-3β-ol |
| M41 | lathosterone | cholest-7-en-3-one |
| M42 | Δ7-dafachronic acid | (25s)-3-oxocholest-7-en-26-oic acid |
| M43 | - | cholest-4-en-3-one |
| M44 | Δ4-dafachronic acid | (25s)-3-oxocholest-7-en-26-oic acid |
| M45 | lophenol | 4α-methyl-cholest-7-en-3β-ol |
| M46 | 4α-methylcholest-8(14)-enol | 4α-methyl-5α-cholest-8(14)-en-3β-ol |
| M47 | 4β -hydroxymethyl-4α -methyl-zymosterol | 4β -hydroxymethyl-4α -methyl-cholesta-8,24-dien-3β-ol |
| M48 | 4β -formyl-4α -methyl-zymosterol | 4β -formyl-4α -methyl-cholesta-8,24-dien-3β-ol |
| M49 | 4β -carboxy-4α -methyl-zymosterol | 4β -carboxy-4α -methyl-cholesta-8,24-dien-3β-ol |
| M50 | - | 4α-methyl-5α-cholesta-8-en-3β-ol |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darnet, S.; Schaller, H. Metabolism and Biological Activities of 4-Methyl-Sterols. Molecules 2019, 24, 451. https://doi.org/10.3390/molecules24030451
Darnet S, Schaller H. Metabolism and Biological Activities of 4-Methyl-Sterols. Molecules. 2019; 24(3):451. https://doi.org/10.3390/molecules24030451
Chicago/Turabian StyleDarnet, Sylvain, and Hubert Schaller. 2019. "Metabolism and Biological Activities of 4-Methyl-Sterols" Molecules 24, no. 3: 451. https://doi.org/10.3390/molecules24030451