Co-Localization of DNA i-Motif-Forming Sequences and 5-Hydroxymethyl-cytosines in Human Embryonic Stem Cells
Abstract
:1. Introduction
2. Results
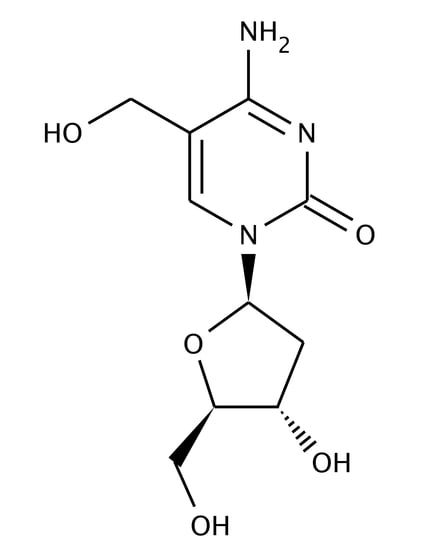
2.1. Very Few iM-Forming Sequences Undergo 5-Hydroxymethylation
2.2. 5hmCs and G4/iMs Distributions are Asymmetric around the TSS
2.3. Putative G4/iMs Enriched with 5hmC are Primarily Found in Differentiation and Proliferation Genes
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Patel, D.J.; Phan, A.T.; Kuryavyi, V. Human telomere, oncogenic promoter and 5 5 gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2007, 35, 7429–7455. [Google Scholar] [CrossRef] [PubMed]
- Catasti, P.; Chen, X.; Deaven, L.L.; Moyzis, R.K.; Bradbury, E.M.; Gupta, G. Cystosine-rich strands of the insulin minisatellite adopt hairpins with intercalated Cytosine+·Cytosine pairs11Edited by I. Tinoco. J. Mol. Biol. 1997, 272, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Hurley, L.H.; Wheelhouse, R.T.; Sun, D.; Kerwin, S.M.; Salazar, M.; Fedoroff, O.Y.; Han, F.X.; Han, H.; Izbicka, E.; Von Hoff, D.D. G-quadruplexes as targets for drug design. Pharmacol. Ther. 2000, 85, 141–158. [Google Scholar] [CrossRef]
- Guéron, M.; Leroy, J.-L. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000, 10, 326–331. [Google Scholar] [CrossRef]
- Han, H.; Hurley, L.H. G-quadruplex DNA: A potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000, 21, 136–141. [Google Scholar] [CrossRef]
- Zhao, Y.; Du, Z.; Li, N. Extensive selection for the enrichment of G4 DNA motifs in transcriptional regulatory regions of warm blooded animals. FEBS Lett. 2007, 581, 1951–1956. [Google Scholar] [CrossRef] [Green Version]
- Cuesta, J.; Read, M.A.; Neidle, S. The Design of G-quadruplex Ligands as Telomerase Inhibitors. Mini Reviews Med. Chem. 2003, 3, 11–21. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Gehring, K.; Leroy, J.L.; Guéron, M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef]
- Bhavsar-Jog, Y.P.; Van Dornshuld, E.; Brooks, T.A.; Tschumper, G.S.; Wadkins, R.M. Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA. Biochemistry 2014, 53, 1586–1594. [Google Scholar] [CrossRef]
- Cui, J.; Waltman, P.; Le, V.H.; Lewis, E.A. The effect of molecular crowding on the stability of human c-MYC promoter sequence i-motif at neutral pH. Molecules 2013, 18, 12751–12767. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Nakano, S.; Sugimoto, N. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 2010, 46, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Reilly, S.M.; Morgan, R.K.; Brooks, T.A.; Wadkins, R.M. Effect of Interior Loop Length on the Thermal Stability and p K a of i-Motif DNA. Biochemistry 2015, 54, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.P.; Waller, Z.A.E.; Huppert, J.L. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017, 45, 2951–2959. [Google Scholar] [CrossRef]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Dzatko, S.; Krafcikova, M.; Hänsel-Hertsch, R.; Fessl, T.; Fiala, R.; Loja, T.; Krafcik, D.; Mergny, J.-L.; Foldynova-Trantirkova, S.; Trantirek, L. Evaluation of the Stability of DNA i-Motifs in the Nuclei of Living Mammalian Cells. Angew. Chem. Int. Ed. 2018, 57, 2165–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; O’Neill, C. Methylation and hydroxymethylation of CpG display dynamic landscapes in early embryo development and define differentiation into embryonic and placental lineages. Epigenetics Chromatin 2013, 6, P60. [Google Scholar] [CrossRef]
- Lister, R.; Ecker, J.R. Finding the fifth base: Genome-wide sequencing of cytosine methylation. Genome Res. 2009, 19, 959–966. [Google Scholar] [CrossRef] [Green Version]
- Münzel, M.; Globisch, D.; Carell, T. 5-Hydroxymethylcytosine, the Sixth Base of the Genome. Angew. Chem. Int. Ed. 2011, 50, 6460–6468. [Google Scholar] [CrossRef]
- Pastor, W.A.; Pape, U.J.; Huang, Y.; Henderson, H.R.; Lister, R.; Ko, M.; McLoughlin, E.M.; Brudno, Y.; Mahapatra, S.; Kapranov, P.; et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011, 473, 394–397. [Google Scholar] [CrossRef] [Green Version]
- Spruijt, C.G.; Gnerlich, F.; Smits, A.H.; Pfaffeneder, T.; Jansen, P.W.T.C.; Bauer, C.; Münzel, M.; Wagner, M.; Müller, M.; Khan, F.; et al. Dynamic Readers for 5-(Hydroxy)Methylcytosine and Its Oxidized Derivatives. Cell 2013, 152, 1146–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Wu, F.; Tan, L.; Kong, L.; Xiong, L.; Deng, J.; Barbera, A.J.; Zheng, L.; Zhang, H.; Huang, S.; et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol. Cell 2011, 42, 451–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Hon, G.C.; Szulwach, K.E.; Song, C.-X.; Zhang, L.; Kim, A.; Li, X.; Dai, Q.; Shen, Y.; Park, B. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012, 149, 1368–1380. [Google Scholar] [CrossRef] [PubMed]
- Denissenko, M.F.; Chen, J.X.; Tang, M.; Pfeifer, G.P. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc. Natl. Acad. Sci. USA 1997, 94, 3893–3898. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cai, X.; Cai, C.-L.; Wang, J.; Zhang, W.; Petersen, B.E.; Yang, F.-C.; Xu, M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 2011, 118, 4509–4518. [Google Scholar] [CrossRef]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129. [Google Scholar] [CrossRef]
- Wu, H.; D’Alessio, A.C.; Ito, S.; Wang, Z.; Cui, K.; Zhao, K.; Sun, Y.E.; Zhang, Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Gene. Dev. 2011, 25, 679–684. [Google Scholar] [CrossRef] [Green Version]
- Morgan, R.K.; Molnar, M.M.; Batra, H.; Summerford, B.; Wadkins, R.M.; Brooks, T.A. Effects of 5-Hydroxymethylcytosine Epigenetic Modification on the Stability and Molecular Recognition of VEGF i-Motif and G-Quadruplex Structures. J. Nucleic Acids 2018, 2018, 14. [Google Scholar] [CrossRef]
- Huppert, J.L. Four-stranded DNA: Cancer, gene regulation and drug development. Philos. Trans. R. Soc. A 2007, 365, 2969–2984. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44. [Google Scholar] [CrossRef]
- Hazel, P.; Huppert, J.; Balasubramanian, S.; Neidle, S. Loop-length dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004, 126, 16405–16415. [Google Scholar] [CrossRef]
- Das, P.; Golde, T. Dysfunction of TGF-β signaling in Alzheimer’s disease. J. Clin. Investig. 2006, 116, 2855–2857. [Google Scholar] [CrossRef] [PubMed]
- Gezen-Ak, D.; Dursun, E.; Ertan, T.; Hanagasi, H.; Gürvit, H.; Emre, M.; Eker, E.; Oztürk, M.; Engin, F.; Yilmazer, S. Association between Vitamin D Receptor Gene Polymorphism and Alzheimer’s Disease. Tohoku J. Exp. Med. 2007, 212, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.J.; Haas, S.; Jurgens, T. On the Physiological Relevance of Muscarinic Acetylcholine Receptors in Alzheimer’s Disease. Curr. Med. Chem. 2005, 12, 2915–2921. [Google Scholar] [CrossRef] [PubMed]
- Shaftel, S.S.; Griffin, W.S.T.; O’Banion, M.K. The role of interleukin-1 in neuroinflammation and Alzheimer disease: An evolving perspective. J. Neuroinflamm. 2008, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Li, X.; Yan, L.; Tan, Y.; Li, R.; Zhao, Y.; Wang, Y.; Xie, J.; Zhang, Y.; Song, C.; et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014, 15, R49. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133. [Google Scholar] [CrossRef] [PubMed]
- Scaria, V.; Hariharan, M.; Arora, A.; Maiti, S. Quadfinder: Server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res. 2006, 34, W683–W685. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [Green Version]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2012, 41, D377–D386. [Google Scholar] [CrossRef]
Sample Availability: Not available. |




| 1 kb upstream of TSS | Correlation | Lower 95% | Upper 95% | Signif. Prob | |
|---|---|---|---|---|---|
| iM Potential | GC content | 0.29 | 0.25 | 0.34 | <0.0001 * |
| 5hmC content | GC content | 0.12 | 0.07 | 0.17 | <0.0001 * |
| 5hmC content | iM Potential | −0.07 | −0.12 | −0.02 | 0.01 * |
| 1 kb downstream of TSS | |||||
| iM Potential | GC content | 0.30 | 0.25 | 0.34 | <0.0001 * |
| 5hmC content | GC content | 0.06 | 0.01 | 0.10 | <0.0293 * |
| 5hmC content | iM Potential | −0.02 | −0.07 | 0.03 | 0.4782 |
| Differentiation Proliferation Apoptosis | Embryogenesis | Transcription Translation | Metabolism/Biosynthesis Cytoskeletal Organization Transport/Ion Binding Enzyme Activity |
|---|---|---|---|
| BIRC7 (5) protein binding, peptidase inhibitor activity | CST3 (4) protein binding | MRPS24 (3) structural constituent of ribosome, nucleic acid binding | BAIAP2L2 (5) receptor activity |
| CHRM1 (4) G-protein coupled receptor activity | DTX1 (3) developmental process | C17orf49 (3) nucleic acid binding | CDIPT (3) transferase activity |
| CST3 (4) protein binding, cysteine-type endopeptidase inhibitor activity | DUSP2 (3) phosphoprotein phosphatase activity, protein binding, kinase inhibitor activity, kinase regulator activity | SORBS3 (3) structural constituent of cytoskeleton | CYP27C1 (3) oxidoreductase activity |
| CYGB (4) blood circulation, transport | FOXH1 (5) transcription factor activity | ZGPAT (3) Negative regulation of transcription | EPS8L2 (3) intracellular signaling cascade, cell motion intracellular signaling cascade |
| DTX1 (3) developmental process | PLEC (4) structural constituent of cytoskeleton, calcium ion binding actin binding | FOXH1 (5) transcription factor activity | ITIH4 (3) protein binding, serine-type endopeptidase inhibitor activity |
| DUSP2 (3) phosphoprotein phosphatase activity, protein binding, kinase inhibitor activity, kinase regulator activity | TNFSF13 (3) cytokine binding, Tumor necrosis factor receptor binding | MAMSTR (4) transcription factor binding, nucleic acid binding | KCTD6 (3) protein binding |
| FOXH1 (5) transcription factor activity | PRR15L (3) Negative regulation of transcription | MPDU1 (3) lipid metabolic process, protein amino acid glycosylation | |
| HSPBP1 (4) protein binding, enzyme regulator activity | PADI3 (3) hydrolase activity | ||
| MAMSTR (4) transcription factor binding nucleic acid binding | |||
| NHP2 (4) structural constituent of ribosome, nucleic acid binding | |||
| PRR15L (3) Negative regulation of transcription | |||
| RASGRP4 (4) calcium ion binding, receptor binding, small GTPase regulator activity, guanyl-nucleotide exchange factor activity |
| Differentiation Proliferation Apoptosis | Embryogenesis | Transcription Translation | Metabolism/Biosynthesis Cytoskeletal Organization Transport/Ion Binding Enzyme Activity |
|---|---|---|---|
| CTCFL (7) transcription factor activity | H1FOO (3) DNA binding | VAV1(3) receptor binding, small GTPase regulator activity, guanyl-nucleotide exchange factor activity | EMILIN1(3) cell adhesion |
| ENG (4) transforming growth factor, beta receptor activity, cytokine receptor activity | HHIPL1(3) G-protein coupled receptor activity | EPS8L1 (4) intracellular signalling cascade cell motion, intracellular signalling cascade | |
| GPR55(3) G-protein coupled receptor activity | SCUBE2 visual perception, sensory perception, signal transduction, cell-cell adhesion, mesoderm development | IP6K3 (6) kinase activity | |
| H1FOO (3) DNA binding | VIL1(3) structural constituent of cytoskeleton, actin binding | MFSD4(3) transport | |
| HHIPL1(3) G-protein coupled receptor activity | NME4 (4) nucleotide kinase activity | ||
| PLCB2(3) phospholipase activity, calcium ion binding, receptor binding, small GTPase regulator activity, guanyl-nucleotide exchange factor activity | S100A16 (3) calcium ion binding, receptor binding calmodulin binding | ||
| SCUBE2(3) visual perception, sensory perception, signal transduction, cell-cell adhesion, mesoderm development | SLC25A23 (3) amino acid transmembrane, transporter activity, transmembrane transporter activity, calcium ion binding, calmodulin binding | ||
| SP100 (3) transcription factor activity chromatin binding, receptor binding, transcription factor activity | SLC37A1 (3) cation transmembrane, transporter activity | ||
| VIL1(3) structural constituent of cytoskeleton, actin binding | TYROBP (3) receptor activity | ||
| VAV1(3) receptor binding, small GTPase regulator activity, guanyl-nucleotide exchange factor activity | DPEP1 Metalo-peptidase activity |
| Biological Processes Cluster 1 | Biological Processes Cluster 2 | |||
|---|---|---|---|---|
| Regulation of Protein Kinase Activity (p = 0.02) | Regulation of Kinase Activity (p = 0.02) | Regulation of Transferase Activity (p = 0.02) | Negative Regulation of Macromolecular Metabolic Processes (p = 0.03) | Negative Regulation of Transcription (p = 0.04) |
| ZGPAT | ZGPAT | ZGPAT | PRR15L | PRR15L |
| BIRC7 | BIRC7 | BIRC7 | CST3 | FOXH1 |
| CHRM1 | CHRM1 | CHRM1 | FOXH1 | SORBS3 |
| DUSP2 | DUSP2 | DUSP2 | SORBS3 | ZGPAT |
| ZGPAT | ||||
| Biological Processes Cluster 1 | Biological Processes Cluster 2 | ||
|---|---|---|---|
| Metal Ion Binding (p = 0.01) | Calcium Ion Binding (p = 0.004) | Identical Protein Binding (p = 0.04) | Protein Homodimerization Activity (p = 0.06) |
| CTCFL | S100A16 | S100A16 | S100A16 |
| S100A16 | PLCB2 | SP100 | SP100 |
| SP100 | SCUBE2 | EMILIN1 | ENG |
| NME4 | SLC25A23 | ENG | |
| PLCB2 | VIL1 | ||
| SCUBE2 | |||
| SLC25A3 | |||
| VAV1 | |||
| VIL1 | |||
| DPEP1 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhavsar-Jog, Y.P.; Van Dornshuld, E.; Brooks, T.A.; Tschumper, G.S.; Wadkins, R.M. Co-Localization of DNA i-Motif-Forming Sequences and 5-Hydroxymethyl-cytosines in Human Embryonic Stem Cells. Molecules 2019, 24, 3619. https://doi.org/10.3390/molecules24193619
Bhavsar-Jog YP, Van Dornshuld E, Brooks TA, Tschumper GS, Wadkins RM. Co-Localization of DNA i-Motif-Forming Sequences and 5-Hydroxymethyl-cytosines in Human Embryonic Stem Cells. Molecules. 2019; 24(19):3619. https://doi.org/10.3390/molecules24193619
Chicago/Turabian StyleBhavsar-Jog, Yogini P., Eric Van Dornshuld, Tracy A. Brooks, Gregory S. Tschumper, and Randy M. Wadkins. 2019. "Co-Localization of DNA i-Motif-Forming Sequences and 5-Hydroxymethyl-cytosines in Human Embryonic Stem Cells" Molecules 24, no. 19: 3619. https://doi.org/10.3390/molecules24193619