1. Introduction
Biocatalysts may represent a green and sustainable alternative to chemical catalysts. They can be used under mild conditions and are biodegradable [
1]. Due to their high selectivity and specificity, biocatalysis is becoming more the method of choice for the industrial preparation of chiral compounds [
2]. Nevertheless, the application of wild type biocatalysts for industrial purposes is often not cost-effective, due to low activities and space-time yields [
1,
3]. This can be circumvented by the usage of recombinant microorganisms and optimized enzymes [
3].
The asymmetric reduction of C=C double bonds is a widely employed reaction, since up to two stereogenic centers can be generated [
4]. In this context, ene reductases (ERs) from the old yellow enzyme family (OYE, EC 1.6.99.1) represent highly interesting biocatalysts for
anti-specific hydrogenations of activated alkenes [
5]. Thus, these enzymes have already been applied in industrial-scale biotransformations [
5]. However, large-scale production processes with OYEs are cost-intensive and only applicable when products of high value are produced, e.g., active pharmaceutical compounds. One reason for this is that the majority of these oxidoreductases prefer Nicotinamide adenine dinucleotide phosphate (NADPH) over the less expensive and more stable Nicotinamide Adenine Dinucleotide (NADH) as a cosubstrate [
6,
7,
8]. This is not only a drawback if the biocatalysts are used as isolated enzymes and the expensive cosubstrate has to be added externally, but also if the cosubstrate is supplied through the application of whole-cell biocatalysts. The greater intracellular concentration of NAD(H), which is, for instance, up to 20-fold higher than the concentration of NADP(H) in exponentially growing
Escherichia coli (
E. coli) cells [
9], makes the utilization of this cosubstrate more effective. Additionally, to further reduce process costs, industrial-scale biotransformations necessitating nicotinamide-based cosubstrates are generally conducted with a regeneration system. Thus, an additional advantage of NAD(H)-usage is represented by a larger variety of NAD(H)-regenerating instead of NADP(H)-regenerating systems [
10], which facilitates their implementation in industrial processes [
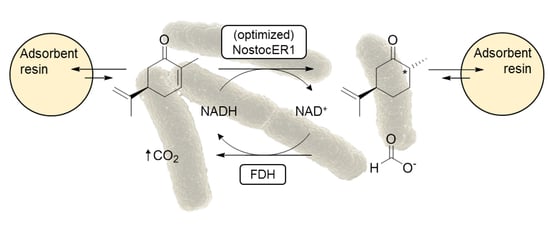
11].
Enzyme engineering was applied to reduce the strong NADPH-dependency of OYEs. In a previous work, we were able to influence the cosubstrate preference of the cyanobacterial ene reductase 1 from
Nostoc sp. PCC7120 (NostocER1) [
12]. This was achieved by swapping loop regions that participate in cosubstrate binding. Different regions of NostocER1 were exchanged for the respective regions of two native donor OYEs with a high activity with NADH or a high affinity toward NADH. Thus, the NADH-binding properties of the donors could be transferred to the cyanobacterial ER. This enabled a combination of the very high stereospecifity and broad substrate spectrum of NostocER1 [
13] with a high NADH activity. Kinetic parameters of NostocER1 and the three best performing NostocER1-variants are listed in
Table 1.
In this study, these optimized enzymes should be applied in a biotransformation of industrial relevance. Therefore, the reduction of (
R)-carvone to (2
R,5
R)-dihydrocarvone was carried out. Dihydrocarvone is used in the synthesis of different molecules of biological interest, e.g., tetraoxane derivatives as compounds with antimalarial activity [
14], keto decalin derivatives as insect antifeedants [
15], or the terpenes (−)-thujopsene [
16] and (+)-decipienin A [
17]. Due to the reduced appearance of undesired side products during the reduction of (
R)-carvone, especially when compared to Baker’s yeast [
18,
19], recombinant
E. coli biocatalysts overexpressing optimized NostocER1 mutants were applied.
3. Discussion
In this study, (
R)-carvone was successfully reduced to (2
R,5
R)-dihydrocarvone using recombinant
E. coli whole-cell biocatalysts overexpressing
nostocER1 variants and
fdh genes. The application of bacterial whole-cell biocatalysts for this reduction reaction was already demonstrated to be beneficial in comparison to other microorganisms, since conversions of ≥96% were reached whereas the undesired reduction of carbonyl bonds by endogenous carbonyl reductases was not observed [
19,
22,
23]. The reduction of the C=O bonds of substrate or product was mostly an issue when applying other organisms like yeasts [
18,
24,
25] and other fungi [
24,
26], plant cells [
27], or microalgae [
28].
In a previous work, the overexpression of the NADPH-preferring cyanobacterial ene reductase from
Nostoc sp. PCC7120 (NostocER1) in combination with a NADP
+-accepting mutant of formate dehydrogenase (FDH
3M) from
Mycobacterium vaccae resulted in highly active
E. coli whole-cell biocatalysts [
19]. 300 mM (
R)-carvone were converted to 290.4 mM (2
R,5
R)-dihydrocarvone in a batch process on a liter-scale after 9 h by applying a biocatalyst concentration of 36 g
CDW L
−1. A space-time yield of 32.3 mmol L
−1 h
−1 was achieved with an initial (2
R,5
R)-dihydrocarvone formation rate of 1.3 mmol h
−1 g
CDW−1. In this study, the NostocER1 wild type was overexpressed in combination with the NAD
+-preferring FDH wild type. Thereby,
E. coli biocatalysts could be provided showing a similar activity compared to literature. On a 0.7 L scale 285.4 mM (2
R,5
R)-dihydrocarvone were formed in a batch process within 8 h with an initial product formation rate of 1.3 ± 0.1 mmol h
−1 g
CDW−1, which results in a space-time yield of 35.7 mmol L
−1 h
−1. Thus, within this process time, a yield of 95.1% was achieved based on 210 mmol applied substrate (
R)-carvone. However, further improved biocatalysts were generated by exchanging the NostocER1 wild type with different NADH-accepting NostocER1-variants (
Table 2). The implementation of the Loop 1,2a NostocER1-variant, which possesses a highly increased catalytic constant with NADH [
12], resulted in significant improvement. Thereby, a yield of 95.7% was achieved after a process time of 5 h. The initial product formation rate was increased 2.1-fold to 2.8 ± 0.1 mmol h
−1 g
CDW−1 and the space-time yield after ≥96% conversion was increased 1.8-fold to 57.4 ± 0.1 mmol L
−1 h
−1 when compared to literature [
19]. This was achieved even though the expression level of this ER was between 24.3 ± 3.6% and 33.4 ± 4.9% lower than the levels of the other biocatalysts. Additionally, the application of the two other NostocER1-variants (Loop 1,5 resp. Loop 1,5,2a) led to higher initial (2
R,5
R)-dihydrocarvone formation rates. However, no further improved space-time yields were observed with these two variants in comparison to the NostocER1 wild type biocatalysts.
The bio-reduction of (
R)-carvone has already been studied using many different microorganisms, including bacteria [
19,
23], yeasts [
18,
24,
25], other fungi [
24,
26], plant cells [
27], or microalgae [
28]. With more than 30 different strains, yeasts and yeast-like fungi are the largest group studied for this reaction [
18,
24,
25]. High substrate conversions of ≥96%, which are a crucial requirement for a cost-effective industrial biotransformation, were only achieved with one strain, i.e.,
Hanseniaspora guilliermondii [
24]. However, only a moderate substrate concentration of 6.9 mM was converted. A significantly reduced conversion (63.6%) was detected after the substrate concentration was increased to 10 mM for the bio-reduction with this organism [
25]. Through the application of the yeast
Saccharomyces cerevisiae and the addition of trehalose to the reaction mixture, the substrate loading could be increased to 16.6 mM and a conversion of 74% was reached [
18].
Besides yeasts, bacteria proved to be beneficial for the selective reduction of (
R)-carvone. In this context,
Pseudomonas putida was used to fully convert 6.9 mM of the substrate with a high stereospecificity within 84 h [
23]. A strong improvement was achieved by the application of
E. coli cells overexpressing the ene reductase NostocER1 and a FDH for cosubstrate regeneration. The high effectiveness of this biocatalyst was demonstrated in a biphasic system with the adsorbent resin XAD4, which results in the, by far, highest product formation rate and space-time yield for the bio-reduction of (
R)-carvone in literature [
19]. Furthermore, a high and stereospecific conversion of 96.8% of the substrate was reached within 9 h of processing time [
19]. In this study, this biocatalyst was further improved by applying NADH-accepting variants of NostocER1 [
12], which results in up to 99.4% conversion of 300 mM (
R)-carvone within 6 h of process time. The product formation rate was increased by a factor of up to 2.1 and the space-time yield by a factor of 1.8. Thereby, the Loop 1,2a NostocER1-variant, i.e., the variant with the highest k
cat with NADH of the three applied NostocER1-variants (
Table 1), revealed the biggest improvement. This is indicative of a stronger influence of k
cat with NADH compared to K
m towards NADH on the productivity of the biocatalyst in the given reaction system. This observation is in accordance with the high NAD(H) concentration in
E. coli, which is up to 20-fold higher compared to NADP(H) in exponentially growing cells [
9]. Through a high cosubstrate concentration, the apparent activity is closer to the maximal turnover number (k
cat), which increases the importance of this parameter.
Another important aspect of this study is that we could not only improve the bioreduction of (R)-carvone, but also demonstrate that cosubstrate engineering of an NADPH-dependent oxidoreductase is useful, even if whole-cell biocatalysts are applied.
4. Materials and Methods
4.1. Chemicals, Enzymes, and Bacterial Strains
(R)-carvone (98%), dihydrocarvone (mixture of isomers 77% (2R,5R), and 20% (2S,5R)) and Amberlite® XAD4 were obtained from Sigma Aldrich (St. Louis, MO, USA). Media components, Kanamycin sulfate, and NAD(H) were purchased from Carl Roth (Karlsruhe, Germany). Enzymes for DNA manipulation were bought from New England Biolabs (Ipswich, MA, USA) and primers were synthesized by Eurofins Genomics (Ebersberg, Germany). E. coli DH5α (Invitrogen, Carlsbad, CA, USA) was used for cloning and E. coli BL21 (DE3) (Novagen, San Diego, CA, USA) for protein expression and biotransformation.
4.2. Cloning
Standard procedures were used for polymerase chain reaction, ligation, transformation, and plasmid and cell preparation as described by Mülhart [
29]. The constructs for co-expression of the NostocER1-variants and FDH based on a pET28a(+) vector (Novagen, San Diego, CA, USA). The vectors expression cassettes were modified with an additional T7 promotor and ribosome binding site (rbs) as described before [
19]. Genes encoding for the NostocER1-variants [
12] and the
fdh gene from
Mycobacterium vaccae (GenBank BAB69476.1) were amplified using the primers listed in
Table 3. For amplification Phusion
® High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA) was applied according to the manufacturer’s protocol. For
nostocER1 and all variants of this gene, the restriction sites NcoI and BamHI were used. Additionally,
fdh NdeI and KpnI was applied.
4.3. Cultivation of E. coli
Biocatalyst production in l-scale was performed in four-fold parallel lab-scale stirred-tank reactors with an initial volume of 0.3 L (DASGIP® Bioblock, DASGIP® reactor with two GL45 sides and overhead propulsion, Eppendorf AG, Hamburg, Germany). Temperature, dissolved oxygen concentration (DO), pH, and gas flow rate were monitored during the production of the whole-cell biocatalysts. The temperature was initially set to 37 °C for the batch phase, which was reduced to 30 °C for the fed-batch phase and was further reduced to 20–23 °C after induction for the enzyme production phase. The pH was controlled to pH 7.0 by 12.5% (v/v) ammonia and 1 M phosphoric acid addition. Solved oxygen availability was achieved in the reactors by the supply of sterile air with a sparger at the bottom and dispersion of the gas bubbles with two six-blade Rushton turbines mounted on the axes at a distance of 25 mm. The DO concentration was controlled to a set point of 20% air saturation by varying the stirrer speed between 500 and 1600 rpm. The air flow was set to 60 L h−1 representing an air flow of at least 3.33 volume per volume of the vessel capacity per minute (vvm). DO sensor calibration was performed prior to inoculation and 100% air saturation was achieved by stripping with air at 37 °C. Afterward, the reactors were stripped with nitrogen gas until 0% air saturation was achieved.
Pre-cultures were produced with
terrific broth (TB) medium containing 12 g L
−1 peptone from casein, 24 g L
−1 yeast extract, 5.1 g L
−1 glycerol, 2.1 g L
−1 KH
2PO
4, 12.6 g L
−1 K
2HPO
4, and 34 mg L
−1 kanamycin. The pH was adjusted to pH 7.2 and 4 mL of the TB medium were inoculated either with a single cell colony or cells from a cryo-stock in a 13 mL single use cultivation tube (Sarstedt, Nümbrecht, Germany). Seed cultures and biocatalyst production in stirred-tank bioreactors were carried out in defined medium, according to Wilms et al. [
20]. Seed cultures with an initial glucose concentration of 5 g L
−1 were grown overnight at 37 °C in shaking flasks without baffles (250 rpm, shaking diameter of 5 cm). Cells from the seed cultures were harvested by centrifugation (3250×
g, 10 min) and the required amount of the cell pellet was resuspended in fresh medium, according to Wilms et al. Afterward the re-suspended cells were used for inoculation of the stirred-tank bioreactors by sterile single-use syringes through a cannula.
Glucose served as a substrate for the initial batch phase at a concentration of 1 g L
−1. Kanamycin was used as a selection marker at a concentration of 34 mg L
−1. For fed-batch cultivations, a growth rate of µ
set = 0.15 h
−1 was set by using a biomass yield coefficient of 0.4 g
CDW g
S−1 and, adding a feeding solution with 100 g L
−1 glucose, 19.8 g L
−1 (NH
4)
2HPO
4 and 34 mg L
−1 kanamycin, according to Sun et al. [
30]. Temperature was reduced to 20 °C after a process time of 19.5 h and 1 h prior to the induction of enzyme production with 200 µM IPTG. 7.5 h after induction, the temperature was raised to 23 °C due to glucose accumulation in the stirred-tank reactor. During the production phase, the growth rate was reduced to µ
set = 0.04 h
−1 and a feeding solution with 500 g L
−1 glucose, 99 g L
−1 (NH
4)
2HPO
4, and 34 mg L
−1 kanamycin was used. Furthermore, 0.5 g L
−1 MgSO
4 · 7 H
2O and 3 mL L
−1 trace element solution [
20] were added to the stirred-tank reactors after 5.5 and 24.5 h. At 24.5 h, an additional 0.1 g L
−1 thiamin was supplemented to the reaction medium. Cells were transferred to centrifugation tubes after cell cultivation and were stored for less than 24 h at 4 °C until further use for preparative biotransformations. The cells for the analysis of the technical replicability were mixed with 12.5% (
w/
v) glycerol and stored at −60 °C.
4.4. Enzyme Activity Assay
Enzyme activities were determined in cell lysates. Therefore, 1 mL E. coli cell suspension was mixed with 0.5 g glass beads (ø 0.25–0.5 mm, Carl Roth, Karlsruhe, Germany) and disrupted for 5 min and 25 s−1 in a Mixer Mill MM 400 (Retsch, Haan, Germany). Debris was separated by centrifugation (17,200× g, 10 min, 4 °C) and the activity in the supernatant was determined photometrically by oxidation (ER) or reduction (FDH) of 500 µM NAD(H) at 340 nm. Assays were conducted in sodium phosphate buffer (100 mM, pH 7.0) at 30 °C using F96 microwell plates (Nunc, Roskilde, Denmark) and a Multiscan™ FC Photometer (Thermo Fisher Scientific, Waltham, MA, USA). Reactions were initiated by the addition of 10 mM maleimide (ER) or 250 mM sodium formate (FDH) as a substrate. Reaction rates were determined by automated linear regression using MATLAB R2015b (The MathWorks, Natick, MA, USA).
4.5. Biotransformations of (R)-Carvone
Biotransformations of (R)-carvone were carried out on two different scales: 15 mL for the evaluation of technical replicability and 0.7 L for the preparative biotransformation in the stirred-tank bioreactors. All biotransformations were performed with 300 mM (R)-carvone and 450 mM sodium formate in 300 mM sodium phosphate buffer (pH 7.0). Amberlite® XAD4 with a wet mass ratio of 3:1 to (R)-carvone was used for in situ substrate feeding and product removal. (R)-carvone and adsorbent resin (harmonic mean size 0.49–0.69 mm, uniformity coefficient ≤2) were incubated for at least 2 h in buffer (50% (v/v) of the total reaction volume) prior to the biotransformations. Reactions were initiated by the addition of the whole-cell biocatalysts resuspended in 300 mM sodium phosphate buffer and the samples were incubated at 25 °C.
On the 15 mL scale, batch biotransformations were performed in glass vials with rolled rim equipped with snap caps (Vmax 20 mL, Carl Roth, Karlsruhe, Germany). Mixing was achieved with x-shaped magnetic stir bars (ø 1 cm, VWR, Darmstadt, Germany) using a multiple stirrer plate Variomag Poly 15 (HP-Labortechnik, Oberschleißheim, Germany) at 900 rpm. Sampling was done with a volume of 300 µL each.
On the 0.7 L scale, batch biotransformations were performed in parallel stirred-tank reactors (DASGIP® Bioblock, DASGIP® reactor with two GL45 sides and overhead propulsion, Eppendorf, Hamburg, Germany). Furthermore, 100 µM NAD+ were added to the reaction mixture. Mixing and dispersion of the gas phase was achieved with two six-bladed Rushton turbines mounted on the axes at a distance of 25 mm and a speed of 700 rpm. The pH was controlled to pH 7.0 by automated addition of 1 M phosphoric acid. Sampling was done with a volume of 1 mL each.
4.6. Analytics
Prior to the analytical procedure, the samples had to be extracted using EtOAc. Therefore, between 33% and 50% (v/v), EtOAc was added to the aqueous-adsorbent resin samples and extraction was performed for 15 min at 25 s−1 in a Mixer Mill MM 400 (Retsch, Haan, Germany) for enhanced mass transfer. The phases were separated by centrifugation (17,200× g, 5 min) and 100 µL of the organic phase was dissolved in 300 µL EtOAc and 100 µL (R)-limonene (36 mM in EtOAc) as an internal standard.
Analytics were performed using a gas chromatograph GC-2010 Plus (Shimadzu, Kyōto, Japan) equipped with a flame ionization detector (FID) and a chiral Lipodex-E column (length 25 m, ø 0.25 mm, Macherey-Nagel, Düren, Germany). The injection volume was set to 1 µL and the split ratio was 10. The injector temperature was set to 200 °C and the detector temperature was set to 250 °C. All runs were performed with the same oven temperature program, i.e., 60 °C for 3 min followed by 3 °C min−1 up to 120 °C. Typical retention times were: (R)-limonene 7.5 min, (2R,5R)-DHC 17.4 min, (2S,5R)-DHC 18.8 min, and (R)-carvone 20.2 min. Compound concentrations were determined using a standard from 0.1 mM to 30 mM of the reference compounds. The application of a biphasic system led to high fluctuations of the determined concentrations. Therefore, substrates and products were summarized and normalized to the applied total concentration of 300 mM. Diastereomeric excesses (de) were determined by comparison of the (2R,5R)-dihydrocarvone concentration with the (2S,5R)-dihydrocarvone concentration. Corresponding standard deviations of three technical replicates were between 0.01% and 0.04%. Initial (2R,5R)-dihydrocarvone formation rates were determined by linear regression until a product concentration of 150 mM was achieved.