Artificial Intelligence (AI) in Biomedical Imaging
Share This Topical Collection
Editor
 Dr. Cristiana Corsi
Dr. Cristiana Corsi
 Dr. Cristiana Corsi
Dr. Cristiana Corsi
E-Mail
Website
Collection Editor
Department of Electrical, Electronics and Information Engineering “Guglielmo Marconi”, Cesena Campus, University of Bologna, Via Venezia 52, 47521 Cesena, Italy
Interests: biomedical image processing; clinical engineering
Topical Collection Information
Dear Colleagues,
We are inviting submissions for an upcoming Topical Collection on AI in biomedical imaging.
Thanks to recent advancements in AI, the field of medical imaging has also developed in many innovative ways. Numerous AI-based tools have been developed to automate medical image analysis and improve automated image interpretation. In particular, deep learning approaches have demonstrated exceptional performance in the screening and diagnosis of many diseases. This field is also becoming increasingly accessible to researchers in medicine and biology who have not traditionally been machine learning practitioners due to the limited availability of software libraries. A further challenge regarding AI‐driven solutions is the development of tools for a personalized disease assessment through deep learning models by taking advantage of their ability to learn patterns and relationships in data and utilize massive volumes of medical images.
The aim of this Topical Collection is to capture recent research and seek contributions of high-quality papers focusing on medical image processing and analysis. Of particular interest are submissions regarding AI-driven computer-aided diagnosis and improvement of automated image interpretation. However, contributions concerning other aspects of medical image processing and analysis (including, but not limited to, image quality improvement, image restoration, image segmentation, image registration and radiomics analysis) are also welcomed.
Dr. Cristiana Corsi
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Sensors is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- Machine learning and artificial intelligence for biomedical applications
- Biomedical image analysis
- Biomedical image processing
- Biomedical image reconstruction
- Sensing and detection for biomedical imaging
- Computer aided diagnosis
- Health informatics
- Prediction of clinical outcomes
- Data-efficient domain adaptation and transfer learning
- Synthetic data generation for healthcare
Published Papers (11 papers)
Open AccessArticle
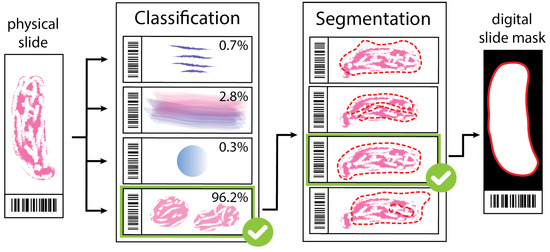
Artifact Augmentation for Enhanced Tissue Detection in Microscope Scanner Systems
by
Dániel Küttel, László Kovács, Ákos Szölgyén, Róbert Paulik, Viktor Jónás, Miklós Kozlovszky and Béla Molnár
Viewed by 905
Abstract
As the field of routine pathology transitions into the digital realm, there is a surging demand for the full automation of microscope scanners, aiming to expedite the process of digitizing tissue samples, and consequently, enhancing the efficiency of case diagnoses. The key to
[...] Read more.
As the field of routine pathology transitions into the digital realm, there is a surging demand for the full automation of microscope scanners, aiming to expedite the process of digitizing tissue samples, and consequently, enhancing the efficiency of case diagnoses. The key to achieving seamless automatic imaging lies in the precise detection and segmentation of tissue sample regions on the glass slides. State-of-the-art approaches for this task lean heavily on deep learning techniques, particularly U-Net convolutional neural networks. However, since samples can be highly diverse and prepared in various ways, it is almost impossible to be fully prepared for and cover every scenario with training data. We propose a data augmentation step that allows artificially modifying the training data by extending some artifact features of the available data to the rest of the dataset. This procedure can be used to generate images that can be considered synthetic. These artifacts could include felt pen markings, speckles of dirt, residual bubbles in covering glue, or stains. The proposed approach achieved a 1–6% improvement for these samples according to the F
1 Score metric.
Full article
►▼
Show Figures
Open AccessArticle
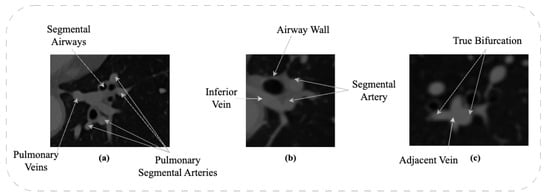
DRU-Net: Pulmonary Artery Segmentation via Dense Residual U-Network with Hybrid Loss Function
by
Manahil Zulfiqar, Maciej Stanuch, Marek Wodzinski and Andrzej Skalski
Cited by 1 | Viewed by 1446
Abstract
The structure and topology of the pulmonary arteries is crucial to understand, plan, and conduct medical treatment in the thorax area. Due to the complex anatomy of the pulmonary vessels, it is not easy to distinguish between the arteries and veins. The pulmonary
[...] Read more.
The structure and topology of the pulmonary arteries is crucial to understand, plan, and conduct medical treatment in the thorax area. Due to the complex anatomy of the pulmonary vessels, it is not easy to distinguish between the arteries and veins. The pulmonary arteries have a complex structure with an irregular shape and adjacent tissues, which makes automatic segmentation a challenging task. A deep neural network is required to segment the topological structure of the pulmonary artery. Therefore, in this study, a Dense Residual U-Net with a hybrid loss function is proposed. The network is trained on augmented Computed Tomography volumes to improve the performance of the network and prevent overfitting. Moreover, the hybrid loss function is implemented to improve the performance of the network. The results show an improvement in the Dice and HD95 scores over state-of-the-art techniques. The average scores achieved for the Dice and HD95 scores are 0.8775 and 4.2624 mm, respectively. The proposed method will support physicians in the challenging task of preoperative planning of thoracic surgery, where the correct assessment of the arteries is crucial.
Full article
►▼
Show Figures
Open AccessArticle
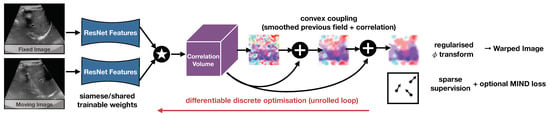
Robust and Realtime Large Deformation Ultrasound Registration Using End-to-End Differentiable Displacement Optimisation
by
Mattias P. Heinrich, Hanna Siebert, Laura Graf, Sven Mischkewitz and Lasse Hansen
Cited by 1 | Viewed by 1397
Abstract
Image registration for temporal ultrasound sequences can be very beneficial for image-guided diagnostics and interventions. Cooperative human–machine systems that enable seamless assistance for both inexperienced and expert users during ultrasound examinations rely on robust, realtime motion estimation. Yet rapid and irregular motion patterns,
[...] Read more.
Image registration for temporal ultrasound sequences can be very beneficial for image-guided diagnostics and interventions. Cooperative human–machine systems that enable seamless assistance for both inexperienced and expert users during ultrasound examinations rely on robust, realtime motion estimation. Yet rapid and irregular motion patterns, varying image contrast and domain shifts in imaging devices pose a severe challenge to conventional realtime registration approaches. While learning-based registration networks have the promise of abstracting relevant features and delivering very fast inference times, they come at the potential risk of limited generalisation and robustness for unseen data; in particular, when trained with limited supervision. In this work, we demonstrate that these issues can be overcome by using end-to-end differentiable displacement optimisation. Our method involves a trainable feature backbone, a correlation layer that evaluates a large range of displacement options simultaneously and a differentiable regularisation module that ensures smooth and plausible deformation. In extensive experiments on public and private ultrasound datasets with very sparse ground truth annotation the method showed better generalisation abilities and overall accuracy than a VoxelMorph network with the same feature backbone, while being two times faster at inference.
Full article
►▼
Show Figures
Open AccessArticle
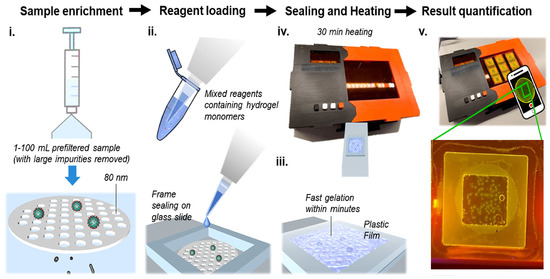
Deep Learning Solution for Quantification of Fluorescence Particles on a Membrane
by
Abdellah Zakaria Sellam, Azeddine Benlamoudi, Clément Antoine Cid, Leopold Dobelle, Amina Slama, Yassin El Hillali and Abdelmalik Taleb-Ahmed
Cited by 1 | Viewed by 1805
Abstract
The detection and quantification of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus particles in ambient waters using a membrane-based in-gel loop-mediated isothermal amplification (mgLAMP) method can play an important role in large-scale environmental surveillance for early warning of potential outbreaks. However, counting particles
[...] Read more.
The detection and quantification of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) virus particles in ambient waters using a membrane-based in-gel loop-mediated isothermal amplification (mgLAMP) method can play an important role in large-scale environmental surveillance for early warning of potential outbreaks. However, counting particles or cells in fluorescence microscopy is an expensive, time-consuming, and tedious task that only highly trained technicians and researchers can perform. Although such objects are generally easy to identify, manually annotating cells is occasionally prone to fatigue errors and arbitrariness due to the operator’s interpretation of borderline cases. In this research, we proposed a method to detect and quantify multiscale and shape variant SARS-CoV-2 fluorescent cells generated using a portable (
mgLAMP) system and captured using a smartphone camera. The proposed method is based on the YOLOv5 algorithm, which uses CSPnet as its backbone. CSPnet is a recently proposed convolutional neural network (CNN) that duplicates gradient information within the network using a combination of Dense nets and ResNet blocks, and bottleneck convolution layers to reduce computation while at the same time maintaining high accuracy. In addition, we apply the test time augmentation (TTA) algorithm in conjunction with YOLO’s one-stage multihead detection heads to detect all cells of varying sizes and shapes. We evaluated the model using a private dataset provided by the Linde + Robinson Laboratory, California Institute of Technology, United States. The model achieved a mAP@0.5 score of 90.3 in the YOLOv5-s6.
Full article
►▼
Show Figures
Open AccessReview
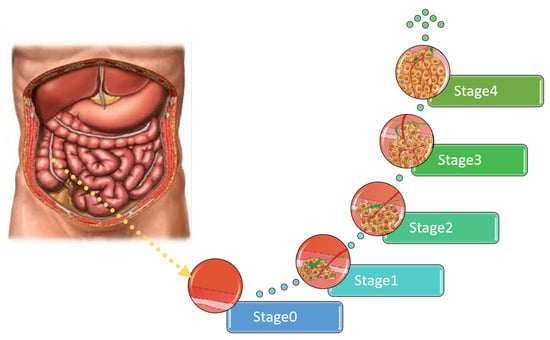
Colon Cancer Diagnosis Based on Machine Learning and Deep Learning: Modalities and Analysis Techniques
by
Mai Tharwat, Nehal A. Sakr, Shaker El-Sappagh, Hassan Soliman, Kyung-Sup Kwak and Mohammed Elmogy
Cited by 5 | Viewed by 6992
Abstract
The treatment and diagnosis of colon cancer are considered to be social and economic challenges due to the high mortality rates. Every year, around the world, almost half a million people contract cancer, including colon cancer. Determining the grade of colon cancer mainly
[...] Read more.
The treatment and diagnosis of colon cancer are considered to be social and economic challenges due to the high mortality rates. Every year, around the world, almost half a million people contract cancer, including colon cancer. Determining the grade of colon cancer mainly depends on analyzing the gland’s structure by tissue region, which has led to the existence of various tests for screening that can be utilized to investigate polyp images and colorectal cancer. This article presents a comprehensive survey on the diagnosis of colon cancer. This covers many aspects related to colon cancer, such as its symptoms and grades as well as the available imaging modalities (particularly, histopathology images used for analysis) in addition to common diagnosis systems. Furthermore, the most widely used datasets and performance evaluation metrics are discussed. We provide a comprehensive review of the current studies on colon cancer, classified into deep-learning (DL) and machine-learning (ML) techniques, and we identify their main strengths and limitations. These techniques provide extensive support for identifying the early stages of cancer that lead to early treatment of the disease and produce a lower mortality rate compared with the rate produced after symptoms develop. In addition, these methods can help to prevent colorectal cancer from progressing through the removal of pre-malignant polyps, which can be achieved using screening tests to make the disease easier to diagnose. Finally, the existing challenges and future research directions that open the way for future work in this field are presented.
Full article
►▼
Show Figures
Open AccessArticle
RDCTrans U-Net: A Hybrid Variable Architecture for Liver CT Image Segmentation
by
Lingyun Li and Hongbing Ma
Cited by 16 | Viewed by 2616
Abstract
Segmenting medical images is a necessary prerequisite for disease diagnosis and treatment planning. Among various medical image segmentation tasks, U-Net-based variants have been widely used in liver tumor segmentation tasks. In view of the highly variable shape and size of tumors, in order
[...] Read more.
Segmenting medical images is a necessary prerequisite for disease diagnosis and treatment planning. Among various medical image segmentation tasks, U-Net-based variants have been widely used in liver tumor segmentation tasks. In view of the highly variable shape and size of tumors, in order to improve the accuracy of segmentation, this paper proposes a U-Net-based hybrid variable structure—RDCTrans U-Net for liver tumor segmentation in computed tomography (CT) examinations. We design a backbone network dominated by ResNeXt50 and supplemented by dilated convolution to increase the network depth, expand the perceptual field, and improve the efficiency of feature extraction without increasing the parameters. At the same time, Transformer is introduced in down-sampling to increase the network’s overall perception and global understanding of the image and to improve the accuracy of liver tumor segmentation. The method proposed in this paper tests the segmentation performance of liver tumors on the LiTS (Liver Tumor Segmentation) dataset. It obtained 89.22% mIoU and 98.91% Acc, for liver and tumor segmentation. The proposed model also achieved 93.38% Dice and 89.87% Dice, respectively. Compared with the original U-Net and the U-Net model that introduces dense connection, attention mechanism, and Transformer, respectively, the method proposed in this paper achieves SOTA (state of art) results.
Full article
►▼
Show Figures
Open AccessArticle
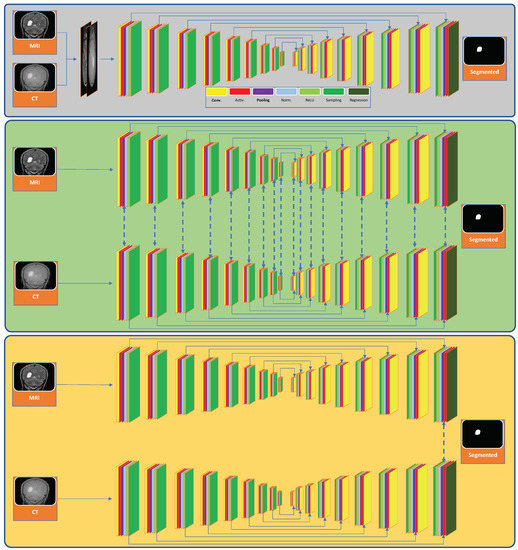
A Deep Learning Framework for Segmenting Brain Tumors Using MRI and Synthetically Generated CT Images
by
Kh Tohidul Islam, Sudanthi Wijewickrema and Stephen O’Leary
Cited by 15 | Viewed by 3847
Abstract
Multi-modal three-dimensional (3-D) image segmentation is used in many medical applications, such as disease diagnosis, treatment planning, and image-guided surgery. Although multi-modal images provide information that no single image modality alone can provide, integrating such information to be used in segmentation is a
[...] Read more.
Multi-modal three-dimensional (3-D) image segmentation is used in many medical applications, such as disease diagnosis, treatment planning, and image-guided surgery. Although multi-modal images provide information that no single image modality alone can provide, integrating such information to be used in segmentation is a challenging task. Numerous methods have been introduced to solve the problem of multi-modal medical image segmentation in recent years. In this paper, we propose a solution for the task of brain tumor segmentation. To this end, we first introduce a method of enhancing an existing magnetic resonance imaging (MRI) dataset by generating synthetic computed tomography (CT) images. Then, we discuss a process of systematic optimization of a convolutional neural network (CNN) architecture that uses this enhanced dataset, in order to customize it for our task. Using publicly available datasets, we show that the proposed method outperforms similar existing methods.
Full article
►▼
Show Figures
Open AccessArticle
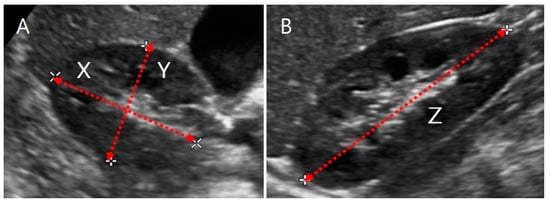
Advanced Kidney Volume Measurement Method Using Ultrasonography with Artificial Intelligence-Based Hybrid Learning in Children
by
Dong-Wook Kim, Hong-Gi Ahn, Jeeyoung Kim, Choon-Sik Yoon, Ji-Hong Kim and Sejung Yang
Cited by 5 | Viewed by 4653
Abstract
In this study, we aimed to develop a new automated method for kidney volume measurement in children using ultrasonography (US) with image pre-processing and hybrid learning and to formulate an equation to calculate the expected kidney volume. The volumes of 282 kidneys (141
[...] Read more.
In this study, we aimed to develop a new automated method for kidney volume measurement in children using ultrasonography (US) with image pre-processing and hybrid learning and to formulate an equation to calculate the expected kidney volume. The volumes of 282 kidneys (141 subjects, <19 years old) with normal function and structure were measured using US. The volumes of 58 kidneys in 29 subjects who underwent US and computed tomography (CT) were determined by image segmentation and compared to those calculated by the conventional ellipsoidal method and CT using intraclass correlation coefficients (ICCs). An expected kidney volume equation was developed using multivariate regression analysis. Manual image segmentation was automated using hybrid learning to calculate the kidney volume. The ICCs for volume determined by image segmentation and ellipsoidal method were significantly different, while that for volume calculated by hybrid learning was significantly higher than that for ellipsoidal method. Volume determined by image segmentation was significantly correlated with weight, body surface area, and height. Expected kidney volume was calculated as (2.22 × weight (kg) + 0.252 × height (cm) + 5.138). This method will be valuable in establishing an age-matched normal kidney growth chart through the accumulation and analysis of large-scale data.
Full article
►▼
Show Figures
Open AccessArticle
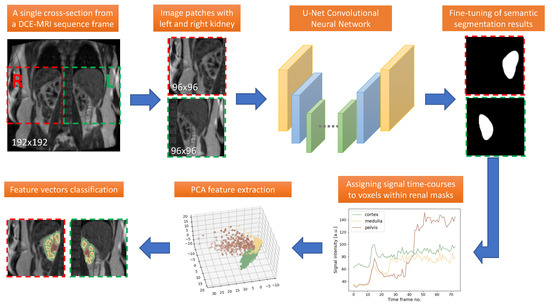
Healthy Kidney Segmentation in the Dce-Mr Images Using a Convolutional Neural Network and Temporal Signal Characteristics
by
Artur Klepaczko, Eli Eikefjord and Arvid Lundervold
Cited by 4 | Viewed by 1989
Abstract
Quantification of renal perfusion based on dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) requires determination of signal intensity time courses in the region of renal parenchyma. Thus, selection of voxels representing the kidney must be accomplished with special care and constitutes one of the
[...] Read more.
Quantification of renal perfusion based on dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) requires determination of signal intensity time courses in the region of renal parenchyma. Thus, selection of voxels representing the kidney must be accomplished with special care and constitutes one of the major technical limitations which hampers wider usage of this technique as a standard clinical routine. Manual segmentation of renal compartments—even if performed by experts—is a common source of decreased repeatability and reproducibility. In this paper, we present a processing framework for the automatic kidney segmentation in DCE-MR images. The framework consists of two stages. Firstly, kidney masks are generated using a convolutional neural network. Then, mask voxels are classified to one of three regions—cortex, medulla, and pelvis–based on DCE-MRI signal intensity time courses. The proposed approach was evaluated on a cohort of 10 healthy volunteers who underwent the DCE-MRI examination. MRI scanning was repeated on two time events within a 10-day interval. For semantic segmentation task we employed a classic U-Net architecture, whereas experiments on voxel classification were performed using three alternative algorithms—support vector machines, logistic regression and extreme gradient boosting trees, among which SVM produced the most accurate results. Both segmentation and classification steps were accomplished by a series of models, each trained separately for a given subject using the data from other participants only. The mean achieved accuracy of the whole kidney segmentation was 94% in terms of IoU coefficient. Cortex, medulla and pelvis were segmented with IoU ranging from 90 to 93% depending on the tissue and body side. The results were also validated by comparing image-derived perfusion parameters with ground truth measurements of glomerular filtration rate (GFR). The repeatability of GFR calculation, as assessed by the coefficient of variation was determined at the level of 14.5 and 17.5% for the left and right kidney, respectively and it improved relative to manual segmentation. Reproduciblity, in turn, was evaluated by measuring agreement between image-derived and iohexol-based GFR values. The estimated absolute mean differences were equal to 9.4 and 12.9 mL/min/1.73 m
2 for scanning sessions 1 and 2 and the proposed automated segmentation method. The result for session 2 was comparable with manual segmentation, whereas for session 1 reproducibility in the automatic pipeline was weaker.
Full article
►▼
Show Figures
Open AccessArticle
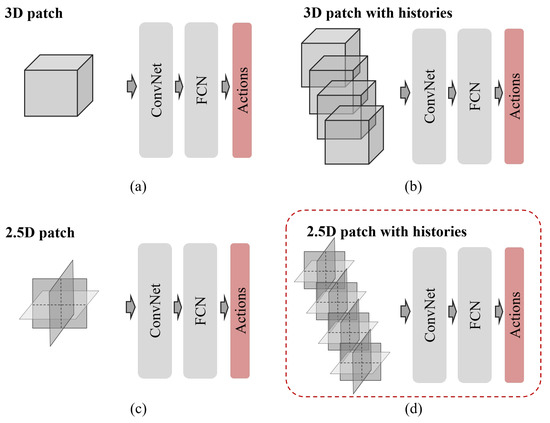
Deep Reinforcement Learning with Explicit Spatio-Sequential Encoding Network for Coronary Ostia Identification in CT Images
by
Yeonggul Jang and Byunghwan Jeon
Cited by 4 | Viewed by 2010
Abstract
Accurate identification of the coronary ostia from 3D coronary computed tomography angiography (CCTA) is a essential prerequisite step for automatically tracking and segmenting three main coronary arteries. In this paper, we propose a novel deep reinforcement learning (DRL) framework to localize the two
[...] Read more.
Accurate identification of the coronary ostia from 3D coronary computed tomography angiography (CCTA) is a essential prerequisite step for automatically tracking and segmenting three main coronary arteries. In this paper, we propose a novel deep reinforcement learning (DRL) framework to localize the two coronary ostia from 3D CCTA. An optimal action policy is determined using a fully explicit spatial-sequential encoding policy network applying 2.5D Markovian states with three past histories. The proposed network is trained using a dueling DRL framework on the CAT08 dataset. The experiment results show that our method is more efficient and accurate than the other methods. blueFloating-point operations (FLOPs) are calculated to measure computational efficiency. The result shows that there are 2.5M FLOPs on the proposed method, which is about 10 times smaller value than 3D box-based methods. In terms of accuracy, the proposed method shows that 2.22 ± 1.12 mm and 1.94 ± 0.83 errors on the left and right coronary ostia, respectively. The proposed method can be applied to the tasks to identify other target objects by changing the target locations in the ground truth data. Further, the proposed method can be utilized as a pre-processing method for coronary artery tracking methods.
Full article
►▼
Show Figures
Open AccessArticle
Automatic Detection of Colorectal Polyps Using Transfer Learning
by
Eva-H. Dulf, Marius Bledea, Teodora Mocan and Lucian Mocan
Cited by 20 | Viewed by 3209
Abstract
Colorectal cancer is the second leading cause of cancer death and ranks third worldwide in diagnosed malignant pathologies (1.36 million new cases annually). An increase in the diversity of treatment options as well as a rising population require novel diagnostic tools. Current diagnostics
[...] Read more.
Colorectal cancer is the second leading cause of cancer death and ranks third worldwide in diagnosed malignant pathologies (1.36 million new cases annually). An increase in the diversity of treatment options as well as a rising population require novel diagnostic tools. Current diagnostics involve critical human thinking, but the decisional process loses accuracy due to the increased number of modulatory factors involved. The proposed computer-aided diagnosis system analyses each colonoscopy and provides predictions that will help the clinician to make the right decisions. Artificial intelligence is included in the system both offline and online image processing tools. Aiming to improve the diagnostic process of colon cancer patients, an application was built that allows the easiest and most intuitive interaction between medical staff and the proposed diagnosis system. The developed tool uses two networks. The first, a convolutional neural network, is capable of classifying eight classes of tissue with a sensitivity of 98.13% and an F1 score of 98.14%, while the second network, based on semantic segmentation, can identify the malignant areas with a Jaccard index of 75.18%. The results could have a direct impact on personalised medicine combining clinical knowledge with the computing power of intelligent algorithms.
Full article
►▼
Show Figures
Planned Papers
The below list represents only planned manuscripts. Some of these
manuscripts have not been received by the Editorial Office yet. Papers
submitted to MDPI journals are subject to peer-review.
Title: Deep Learning-Based Noise Reduction Methods for Low-dose CT Imaging: Comparative Analysis of The Perception-Distortion Tradeoff
Authors: Wonjin KIM; Jang-Hwan CHOI
Affiliation: Ewha Womans University
Abstract: X-ray computed tomography (CT) is widely used in many industries and is an essential tool for clinical diagnosis. Meanwhile it provides a non-invasive way of getting clinical information of patients, high exposure of radiation to patients is another concern in use of CT: increase of use of CT scans can be contributed to potential risk of lung cancer. This makes low-dose CT (LDCT) adopted more than in the past. However, reducing radiation of CT produces more noises in CT scans, thus, research of LDCT denoising is widely conducted in medical imaging fields. Deep neural networks, in recent years, have been successful in almost every field, even solving the most complex problem statements. Research of LDCT denoising has also benefit from this trend and has shown great improvement to reduce noise. This paper investigates about deep learning based LDCT denoisng which are being actively studied in recent years. First, we aim to provide explanations of fundamentals of deep learning based LDCT denoising. Then we provide a comprehensive survey on the recent progress of deep learning based denoising. In general, we conducted a systematic review through to determine the level of evidence of methods for deep learning based LDCT denoising. Additionally, we systematically analyze the research status of LDCT denoising. Finally, we discuss the potentials and open challenge of existing methods and prospect the future directions of deep learning based LDCT denoising.