Crop Genomics and Breeding
A topical collection in Plants (ISSN 2223-7747). This collection belongs to the section "Plant Molecular Biology".
Viewed by 38306Editors
Interests: structural genomics; comparative genomics; evolution; breeding; poaceae
Interests: soybean; genomics; epigenomics
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
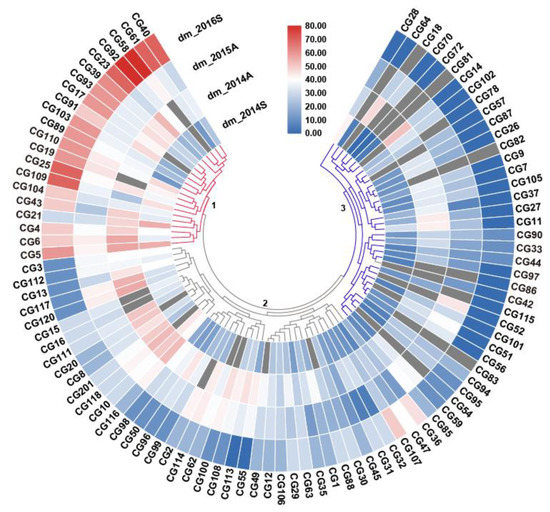
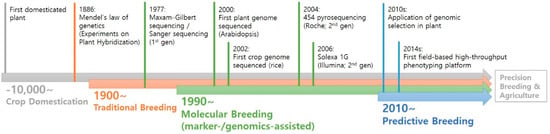
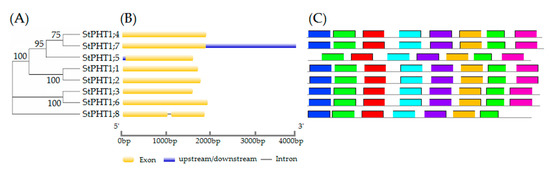
With the advent of next-generation sequencing technology, genomics has been receiving intensive attention for the last two decades. In fact, since the Arabidopsis genome was completed, draft sequences of pseudomolecules have been published for more than 100 plant genomes, including green algae, in large part due to advances in sequencing technologies. Advanced DNA sequencing technologies have also conferred new opportunities for high-throughput low-cost crop genotyping, based on single-nucleotide polymorphisms (SNPs). However, the recurring complication in crop genotyping that differs from other taxa is a high level of DNA sequence duplication, noting that all angiosperms are thought to have polyploidy in their evolutionary history. Nonetheless, advanced genomics technologies have facilitated new opportunities for breeders with a cost-effective, genome-wide scanning, and multiplexed sequencing platform, being able to contribute to many crop breeding programs.
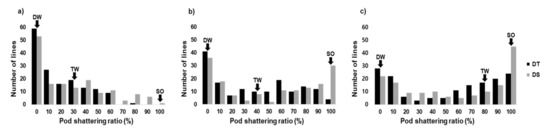
One of the global key phrases in the 21st century is feeding the world. However, foreseeable climate changes can dramatically reduce agricultural productivity, mainly due to abiotic and biotic stresses. To meet the global food demand of an increasing population, food production has to be increased by 60% by 2050. With limited agricultural land, innovative crop cultivation together with the breeding of improved cultivars may be required for resolving the largest part of food security issues.
Plant breeding generally requires the analysis of the genetic background of a target species. In conventional breeding, farmers or breeders traditionally utilized phenotypic markers for selecting desirable plants; however, the use of molecular markers has dramatically changed the landscape of plant breeding. Molecular markers have a number of advantages in plant science in that they are (1) not subject to environmental influence, (2) theoretically available in vast numbers, and (3) usually more objective than phenotypic markers. Consequently, molecular markers can be used for marker-assisted selection (MAS) to greatly increase the efficiency and precision of plant breeding compared to conventional methods. In particular, a plethora of genome data has facilitated the application of sequence-based markers such as simple sequence repeats (SSRs) and SNPs. The frequent use of genomic resources for plant breeding has led to the creation of a new term, genomics-assisted breeding (GAB), integrating all genomics tools and resources. In addition, we are facing a new breeding paradigm, genome selection (GS), by virtue of overflowing genomic and phenotypic data.
With this background, we welcome eminent researchers working on crop genomics across the world to contribute their high-quality original research manuscripts, critical reviews, and opinion articles covering all modern crop genomics and breeding:
- Plant research focusing on food crops including structural, functional, comparative, and evolutionary genomics;
- Application of new genotyping tools;
- Application of genomic tools in plant breeding;
- Genome-wide association study (GWAS) and genetic dissection of complex traits;
- Genome selection and predictive breeding;
- Integration of multi-omics data for prediction of plant phenotypes;
- New breeding techniques for introduction of novel alleles;
- Plant epigenomics research and its potential application in crop improvement.
Prof. Dr. Changsoo Kim
Prof. Dr. Kyung Do Kim
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Plants is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- plant genome
- genomics-assisted breeding
- genome prediction
- sequencing
- bioinformatics