Application of AI in Plants
(Closed)
A topical collection in Plants (ISSN 2223-7747). This collection belongs to the section "Plant Modeling".
Viewed by 54301
Share This Topical Collection
Editors
 Dr. Laura-Jayne Gardiner
Dr. Laura-Jayne Gardiner
 Dr. Laura-Jayne Gardiner
Dr. Laura-Jayne Gardiner
E-Mail
Website
Collection Editor
IBM Research, The Hartree Centre, Warrington WA4 4AD, UK
Interests: genetics; omics; computational biology; AI; polyploid; mapping-by-sequencing; crops and model organisms
 Dr. Ritesh Krishna
Dr. Ritesh Krishna
 Dr. Ritesh Krishna
Dr. Ritesh Krishna
E-Mail
Website
Collection Editor
IBM Research, The Hartree Centre, Warrington WA4 4AD, UK
Interests: bioinformatics; genomics; proteomics; network biology; machine learning; big data; cloud; HPC
Topical Collection Information
Dear Colleagues,
It is predicted that 60% more food will be needed by 2050 in order to feed the increasing global population, while climate change threatens to reduce not only crop yields, but also their nutritious value through mechanisms such as disease, drought, floods, heat, or low nutrient availability. To keep up with the increase in the human population and with environmental changes, it is vital to increase and sustain crop yields, while considering increased pressure to reduce the use of fertilizers, herbicides, insecticides, and fungicides.
Now, more than ever, it is important to provide breeders with new tools that allow them to develop the next generation of crop varieties. Such tools can take various forms, including genetic marker combinations, gene editing strategies, monitoring and management of pests/disease, fertilization schemes, real-time crop imaging/surveillance, and even robotics. The application of artificial intelligence (AI) could be a powerful facilitator towards such goals, particularly in combination with the emerging technologies that are becoming widely adopted in the field, e.g., high-throughput genotyping, multi-omic sequencing approaches, targeted mutagenesis including genome editing, high-throughput phenotyping, and drone imaging.
The objective of this Topical Collection is to show how AI and machine learning (ML) algorithms are being applied to advance plant health and development, and how they could be used and improved in the future in order to aid food security, particularly when this involves the integration of state-of-the-art technologies, e.g., multi-omic datasets. We hope to increase the recognition, as well as the accessibility of AI/ML tools in agricultural and plant research more generally. Review articles on the latest technologies, methodologies, and bottlenecks in the area will also be of interest.
Dr. Laura-Jayne Gardiner
Dr. Ritesh Krishna
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Plants is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- crop improvement
- multi-omics/genotyping
- genetics
- breeding
- biotechnology
- gene editing
- monitoring and surveillance
- phenotyping
- precision agriculture
Published Papers (22 papers)
Open AccessArticle
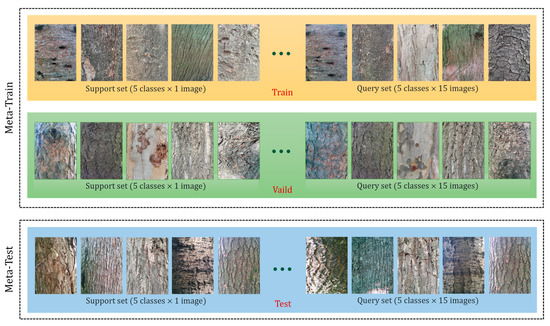
Plant and Disease Recognition Based on PMF Pipeline Domain Adaptation Method: Using Bark Images as Meta-Dataset
by
Zhelin Cui, Kanglong Li, Chunyan Kang, Yi Wu, Tao Li and Mingyang Li
Cited by 1 | Viewed by 1008
Abstract
Efficient image recognition is important in crop and forest management. However, it faces many challenges, such as the large number of plant species and diseases, the variability of plant appearance, and the scarcity of labeled data for training. To address this issue, we
[...] Read more.
Efficient image recognition is important in crop and forest management. However, it faces many challenges, such as the large number of plant species and diseases, the variability of plant appearance, and the scarcity of labeled data for training. To address this issue, we modified a SOTA Cross-Domain Few-shot Learning (CDFSL) method based on prototypical networks and attention mechanisms. We employed attention mechanisms to perform feature extraction and prototype generation by focusing on the most relevant parts of the images, then used prototypical networks to learn the prototype of each category and classify new instances. Finally, we demonstrated the effectiveness of the modified CDFSL method on several plant and disease recognition datasets. The results showed that the modified pipeline was able to recognize several cross-domain datasets using generic representations, and achieved up to 96.95% and 94.07% classification accuracy on datasets with the same and different domains, respectively. In addition, we visualized the experimental results, demonstrating the model’s stable transfer capability between datasets and the model’s high visual correlation with plant and disease biological characteristics. Moreover, by extending the classes of different semantics within the training dataset, our model can be generalized to other domains, which implies broad applicability.
Full article
►▼
Show Figures
Open AccessArticle
MFBP-UNet: A Network for Pear Leaf Disease Segmentation in Natural Agricultural Environments
by
Haoyu Wang, Jie Ding, Sifan He, Cheng Feng, Cheng Zhang, Guohua Fan, Yunzhi Wu and Youhua Zhang
Cited by 4 | Viewed by 1394
Abstract
The accurate prevention and control of pear tree diseases, especially the precise segmentation of leaf diseases, poses a serious challenge to fruit farmers globally. Given the possibility of disease areas being minute with ambiguous boundaries, accurate segmentation becomes difficult. In this study, we
[...] Read more.
The accurate prevention and control of pear tree diseases, especially the precise segmentation of leaf diseases, poses a serious challenge to fruit farmers globally. Given the possibility of disease areas being minute with ambiguous boundaries, accurate segmentation becomes difficult. In this study, we propose a pear leaf disease segmentation model named MFBP-UNet. It is based on the UNet network architecture and integrates a Multi-scale Feature Extraction (MFE) module and a Tokenized Multilayer Perceptron (BATok-MLP) module with dynamic sparse attention. The MFE enhances the extraction of detail and semantic features, while the BATok-MLP successfully fuses regional and global attention, striking an effective balance in the extraction capabilities of both global and local information. Additionally, we pioneered the use of a diffusion model for data augmentation. By integrating and analyzing different augmentation methods, we further improved the model’s training accuracy and robustness. Experimental results reveal that, compared to other segmentation networks, MFBP-UNet shows a significant improvement across all performance metrics. Specifically, MFBP-UNet achieves scores of 86.15%, 93.53%, 90.89%, and 0.922 on MIoU, MP, MPA, and Dice metrics, marking respective improvements of 5.75%, 5.79%, 1.08%, and 0.074 over the UNet model. These results demonstrate the MFBP-UNet model’s superior performance and generalization capabilities in pear leaf disease segmentation and its inherent potential to address analogous challenges in natural environment segmentation tasks.
Full article
►▼
Show Figures
Open AccessArticle
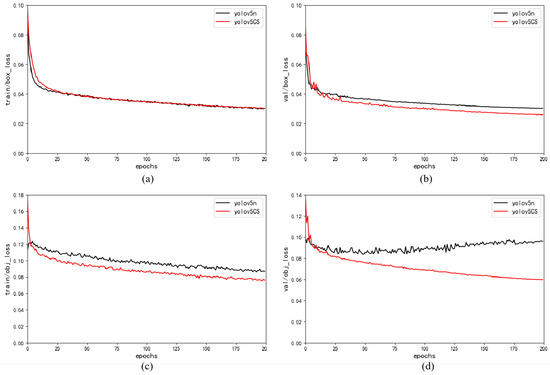
Lightweight Algorithm for Apple Detection Based on an Improved YOLOv5 Model
by
Yu Sun, Dongwei Zhang, Xindong Guo and Hua Yang
Cited by 4 | Viewed by 1539
Abstract
The detection algorithm of the apple-picking robot contains a complex network structure and huge parameter volume, which seriously limits the inference speed. To enable automatic apple picking in complex unstructured environments based on embedded platforms, we propose a lightweight YOLOv5-CS model for apple
[...] Read more.
The detection algorithm of the apple-picking robot contains a complex network structure and huge parameter volume, which seriously limits the inference speed. To enable automatic apple picking in complex unstructured environments based on embedded platforms, we propose a lightweight YOLOv5-CS model for apple detection based on YOLOv5n. Firstly, we introduced the lightweight C3-light module to replace C3 to enhance the extraction of spatial features and boots the running speed. Then, we incorporated SimAM, a parameter-free attention module, into the neck layer to improve the model’s accuracy. The results showed that the size and inference speed of YOLOv5-CS were 6.25 MB and 0.014 s, which were
and 1.2 times that of the YOLOv5n model, respectively. The number of floating-point operations (FLOPs) were reduced by 15.56%, and the average precision (AP) reached 99.1%. Finally, we conducted extensive experiments, and the results showed that the YOLOv5-CS outperformed mainstream networks in terms of AP, speed, and model size. Thus, our real-time YOLOv5-CS model detects apples in complex orchard environments efficiently and provides technical support for visual recognition systems for intelligent apple-picking devices.
Full article
►▼
Show Figures
Open AccessArticle
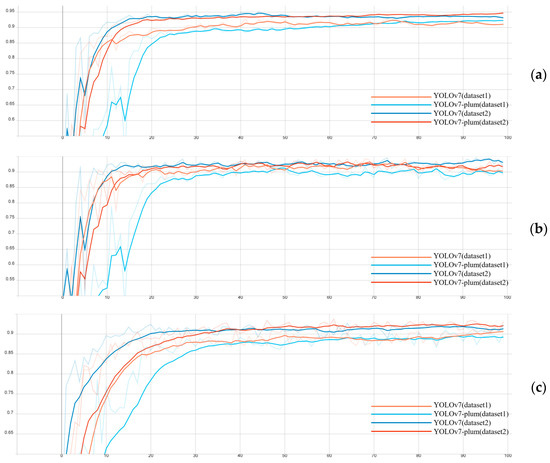
YOLOv7-Plum: Advancing Plum Fruit Detection in Natural Environments with Deep Learning
by
Rong Tang, Yujie Lei, Beisiqi Luo, Junbo Zhang and Jiong Mu
Cited by 5 | Viewed by 1777
Abstract
The plum is a kind of delicious and common fruit with high edible value and nutritional value. The accurate and effective detection of plum fruit is the key to fruit number counting and pest and disease early warning. However, the actual plum orchard
[...] Read more.
The plum is a kind of delicious and common fruit with high edible value and nutritional value. The accurate and effective detection of plum fruit is the key to fruit number counting and pest and disease early warning. However, the actual plum orchard environment is complex, and the detection of plum fruits has many problems, such as leaf shading and fruit overlapping. The traditional method of manually estimating the number of fruits and the presence of pests and diseases used in the plum growing industry has disadvantages, such as low efficiency, a high cost, and low accuracy. To detect plum fruits quickly and accurately in a complex orchard environment, this paper proposes an efficient plum fruit detection model based on an improved You Only Look Once version 7(YOLOv7). First, different devices were used to capture high-resolution images of plum fruits growing under natural conditions in a plum orchard in Gulin County, Sichuan Province, and a dataset for plum fruit detection was formed after the manual screening, data enhancement, and annotation. Based on the dataset, this paper chose YOLOv7 as the base model, introduced the Convolutional Block Attention Module (CBAM) attention mechanism in YOLOv7, used Cross Stage Partial Spatial Pyramid Pooling–Fast (CSPSPPF) instead of Cross Stage Partial Spatial Pyramid Pooling(CSPSPP) in the network, and used bilinear interpolation to replace the nearest neighbor interpolation in the original network upsampling module to form the improved target detection algorithm YOLOv7-plum. The tested YOLOv7-plum model achieved an average precision (AP) value of 94.91%, which was a 2.03% improvement compared to the YOLOv7 model. In order to verify the effectiveness of the YOLOv7-plum algorithm, this paper evaluated the performance of the algorithm through ablation experiments, statistical analysis, etc. The experimental results showed that the method proposed in this study could better achieve plum fruit detection in complex backgrounds, which helped to promote the development of intelligent cultivation in the plum industry.
Full article
►▼
Show Figures
Open AccessArticle
HSSNet: A End-to-End Network for Detecting Tiny Targets of Apple Leaf Diseases in Complex Backgrounds
by
Xing Gao, Zhiwen Tang, Yubao Deng, Shipeng Hu, Hongmin Zhao and Guoxiong Zhou
Cited by 1 | Viewed by 1167
Abstract
Apple leaf diseases are one of the most important factors that reduce apple quality and yield. The object detection technology based on deep learning can detect diseases in a timely manner and help automate disease control, thereby reducing economic losses. In the natural
[...] Read more.
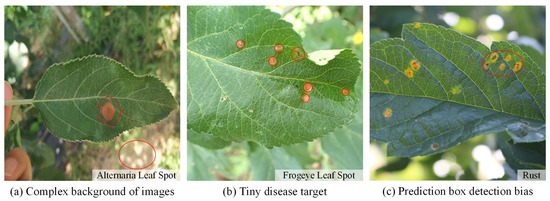
Apple leaf diseases are one of the most important factors that reduce apple quality and yield. The object detection technology based on deep learning can detect diseases in a timely manner and help automate disease control, thereby reducing economic losses. In the natural environment, tiny apple leaf disease targets (a resolution is less than 32 × 32 pixel
2) are easily overlooked. To address the problems of complex background interference, difficult detection of tiny targets and biased detection of prediction boxes that exist in standard detectors, in this paper, we constructed a tiny target dataset TTALDD-4 containing four types of diseases, which include Alternaria leaf spot, Frogeye leaf spot, Grey spot and Rust, and proposed the HSSNet detector based on the YOLOv7-tiny benchmark for professional detection of apple leaf disease tiny targets. Firstly, the H-SimAM attention mechanism is proposed to focus on the foreground lesions in the complex background of the image. Secondly, SP-BiFormer Block is proposed to enhance the ability of the model to perceive tiny targets of leaf diseases. Finally, we use the SIOU loss to improve the case of prediction box bias. The experimental results show that HSSNet achieves 85.04% mAP (mean average precision), 67.53% AR (average recall), and 83 FPS (frames per second). Compared with other standard detectors, HSSNet maintains high real-time detection speed with higher detection accuracy. This provides a reference for the automated control of apple leaf diseases.
Full article
►▼
Show Figures
Open AccessArticle
Classification of Camellia oleifera Diseases in Complex Environments by Attention and Multi-Dimensional Feature Fusion Neural Network
by
Yixin Chen, Xiyun Wang, Zhibo Chen, Kang Wang, Ye Sun, Jiarong Jiang and Xuhao Liu
Viewed by 910
Abstract
The use of neural networks for plant disease identification is a hot topic of current research. However, unlike the classification of ordinary objects, the features of plant diseases frequently vary, resulting in substantial intra-class variation; in addition, the complex environmental noise makes it
[...] Read more.
The use of neural networks for plant disease identification is a hot topic of current research. However, unlike the classification of ordinary objects, the features of plant diseases frequently vary, resulting in substantial intra-class variation; in addition, the complex environmental noise makes it more challenging for the model to categorize the diseases. In this paper, an attention and multidimensional feature fusion neural network (AMDFNet) is proposed for
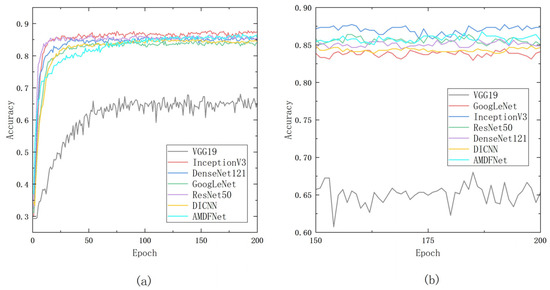
Camellia oleifera disease classification network based on multidimensional feature fusion and attentional mechanism, which improves the classification ability of the model by fusing features to each layer of the Inception structure and enhancing the fused features with attentional enhancement. The model was compared with the classical convolutional neural networks GoogLeNet, Inception V3, ResNet50, and DenseNet121 and the latest disease image classification network DICNN in a self-built camellia disease dataset. The experimental results show that the recognition accuracy of the new model reaches 86.78% under the same experimental conditions, which is 2.3% higher than that of GoogLeNet with a simple Inception structure, and the number of parameters is reduced to one-fourth compared to large models such as ResNet50. The method proposed in this paper can be run on mobile with higher identification accuracy and a smaller model parameter number.
Full article
►▼
Show Figures
Open AccessArticle
Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer
by
Chin Poo Lee, Kian Ming Lim, Yu Xuan Song and Ali Alqahtani
Cited by 8 | Viewed by 3756
Abstract
Plant leaf classification involves identifying and categorizing plant species based on leaf characteristics, such as patterns, shapes, textures, and veins. In recent years, research has been conducted to improve the accuracy of plant classification using machine learning techniques. This involves training models on
[...] Read more.
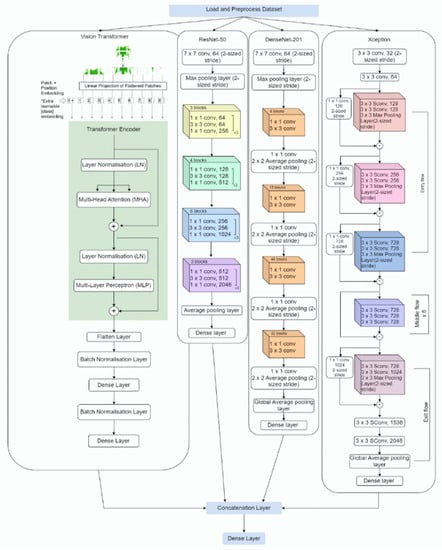
Plant leaf classification involves identifying and categorizing plant species based on leaf characteristics, such as patterns, shapes, textures, and veins. In recent years, research has been conducted to improve the accuracy of plant classification using machine learning techniques. This involves training models on large datasets of plant images and using them to identify different plant species. However, these models are limited by their reliance on large amounts of training data, which can be difficult to obtain for many plant species. To overcome this challenge, this paper proposes a Plant-CNN-ViT ensemble model that combines the strengths of four pre-trained models: Vision Transformer, ResNet-50, DenseNet-201, and Xception. Vision Transformer utilizes self-attention to capture dependencies and focus on important leaf features. ResNet-50 introduces residual connections, aiding in efficient training and hierarchical feature extraction. DenseNet-201 employs dense connections, facilitating information flow and capturing intricate leaf patterns. Xception uses separable convolutions, reducing the computational cost while capturing fine-grained details in leaf images. The proposed Plant-CNN-ViT was evaluated on four plant leaf datasets and achieved remarkable accuracy of 100.00%, 100.00%, 100.00%, and 99.83% on the Flavia dataset, Folio Leaf dataset, Swedish Leaf dataset, and MalayaKew Leaf dataset, respectively.
Full article
►▼
Show Figures
Open AccessArticle
Identification of Soybean Mutant Lines Based on Dual-Branch CNN Model Fusion Framework Utilizing Images from Different Organs
by
Guangxia Wu, Lin Fei, Limiao Deng, Haoyan Yang, Meng Han, Zhongzhi Han and Longgang Zhao
Cited by 1 | Viewed by 1068
Abstract
The accurate identification and classification of soybean mutant lines is essential for developing new plant varieties through mutation breeding. However, most existing studies have focused on the classification of soybean varieties. Distinguishing mutant lines solely by their seeds can be challenging due to
[...] Read more.
The accurate identification and classification of soybean mutant lines is essential for developing new plant varieties through mutation breeding. However, most existing studies have focused on the classification of soybean varieties. Distinguishing mutant lines solely by their seeds can be challenging due to their high genetic similarities. Therefore, in this paper, we designed a dual-branch convolutional neural network (CNN) composed of two identical single CNNs to fuse the image features of pods and seeds together to solve the soybean mutant line classification problem. Four single CNNs (AlexNet, GoogLeNet, ResNet18, and ResNet50) were used to extract features, and the output features were fused and input into the classifier for classification. The results demonstrate that dual-branch CNNs outperform single CNNs, with the dual-ResNet50 fusion framework achieving a 90.22 ± 0.19% classification rate. We also identified the most similar mutant lines and genetic relationships between certain soybean lines using a clustering tree and t-distributed stochastic neighbor embedding algorithm. Our study represents one of the primary efforts to combine various organs for the identification of soybean mutant lines. The findings of this investigation provide a new path to select potential lines for soybean mutation breeding and signify a meaningful advancement in the propagation of soybean mutant line recognition technology.
Full article
►▼
Show Figures
Open AccessArticle
An Accurate Classification of Rice Diseases Based on ICAI-V4
by
Nanxin Zeng, Gufeng Gong, Guoxiong Zhou and Can Hu
Cited by 10 | Viewed by 2293
Abstract
Rice is a crucial food crop, but it is frequently affected by diseases during its growth process. Some of the most common diseases include rice blast, flax leaf spot, and bacterial blight. These diseases are widespread, highly infectious, and cause significant damage, posing
[...] Read more.
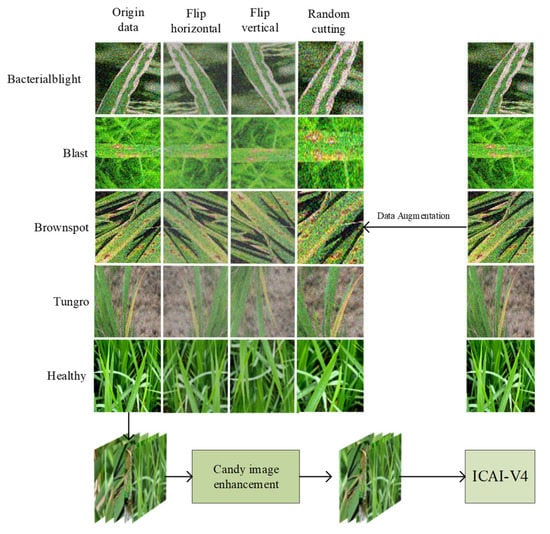
Rice is a crucial food crop, but it is frequently affected by diseases during its growth process. Some of the most common diseases include rice blast, flax leaf spot, and bacterial blight. These diseases are widespread, highly infectious, and cause significant damage, posing a major challenge to agricultural development. The main problems in rice disease classification are as follows: (1) The images of rice diseases that were collected contain noise and blurred edges, which can hinder the network’s ability to accurately extract features of the diseases. (2) The classification of disease images is a challenging task due to the high intra-class diversity and inter-class similarity of rice leaf diseases. This paper proposes the Candy algorithm, an image enhancement technique that utilizes improved Canny operator filtering (the gravitational edge detection algorithm) to emphasize the edge features of rice images and minimize the noise present in the images. Additionally, a new neural network (ICAI-V4) is designed based on the Inception-V4 backbone structure, with a coordinate attention mechanism added to enhance feature capture and overall model performance. The INCV backbone structure incorporates Inception-iv and Reduction-iv structures, with the addition of involution to enhance the network’s feature extraction capabilities from a channel perspective. This enables the network to better classify similar images of rice diseases. To address the issue of neuron death caused by the ReLU activation function and improve model robustness, Leaky ReLU is utilized. Our experiments, conducted using the 10-fold cross-validation method and 10,241 images, show that ICAI-V4 has an average classification accuracy of 95.57%. These results indicate the method’s strong performance and feasibility for rice disease classification in real-life scenarios.
Full article
►▼
Show Figures
Open AccessArticle
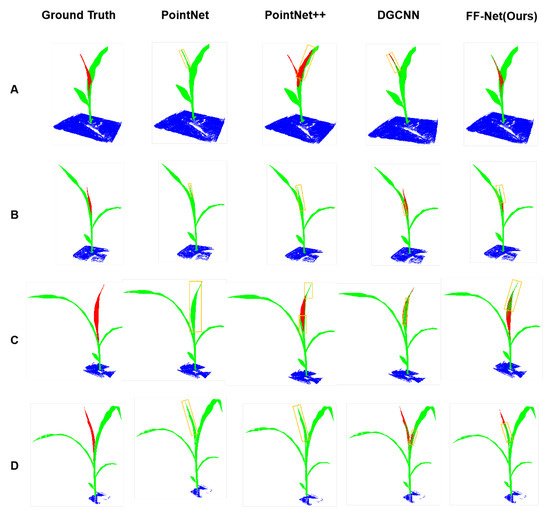
FF-Net: Feature-Fusion-Based Network for Semantic Segmentation of 3D Plant Point Cloud
by
Xindong Guo, Yu Sun and Hua Yang
Cited by 3 | Viewed by 1742
Abstract
Semantic segmentation of 3D point clouds has played an important role in the field of plant phenotyping in recent years. However, existing methods need to down-sample the point cloud to a relatively small size when processing large-scale plant point clouds, which contain more
[...] Read more.
Semantic segmentation of 3D point clouds has played an important role in the field of plant phenotyping in recent years. However, existing methods need to down-sample the point cloud to a relatively small size when processing large-scale plant point clouds, which contain more than hundreds of thousands of points, which fails to take full advantage of the high-resolution of advanced scanning devices. To address this issue, we propose a feature-fusion-based method called FF-Net, which consists of two branches, namely the voxel-branch and the point-branch. In particular, the voxel-branch partitions a point cloud into voxels and then employs sparse 3D convolution to learn the context features, and the point-branch learns the point features within a voxel to preserve the detailed point information. Finally, an attention-based module was designed to fuse the two branch features to produce the final segmentation. We conducted extensive experiments on two large plant point clouds (maize and tomato), and the results showed that our method outperformed three commonly used models on both datasets and achieved the best mIoU of 80.95% on the maize dataset and 86.65% on the tomato dataset. Extensive cross-validation experiments were performed to evaluate the generalization ability of the models, and our method achieved promising segmentation results. In addition, the drawbacks of the proposed method were analyzed, and the directions for future works are given.
Full article
►▼
Show Figures
Open AccessArticle
Study on the Detection Method for Daylily Based on YOLOv5 under Complex Field Environments
by
Hongwen Yan, Songrui Cai, Qiangsheng Li, Feng Tian, Sitong Kan and Meimeng Wang
Cited by 3 | Viewed by 1306
Abstract
Intelligent detection is vital for achieving the intelligent picking operation of daylily, but complex field environments pose challenges due to branch occlusion, overlapping plants, and uneven lighting. To address these challenges, this study selected an intelligent detection model based on YOLOv5s for daylily,
[...] Read more.
Intelligent detection is vital for achieving the intelligent picking operation of daylily, but complex field environments pose challenges due to branch occlusion, overlapping plants, and uneven lighting. To address these challenges, this study selected an intelligent detection model based on YOLOv5s for daylily, the depth and width parameters of the YOLOv5s network were optimized, with Ghost, Transformer, and MobileNetv3 lightweight networks used to optimize the CSPDarknet backbone network of YOLOv5s, continuously improving the model’s performance. The experimental results show that the original YOLOv5s model increased mean average precision (mAP) by 49%, 44%, and 24.9% compared to YOLOv4, SSD, and Faster R-CNN models, optimizing the depth and width parameters of the network increased the mAP of the original YOLOv5s model by 7.7%, and the YOLOv5s model with Transformer as the backbone network increased the mAP by 0.2% and the inference speed by 69% compared to the model after network parameter optimization. The optimized YOLOv5s model provided precision, recall rate, mAP, and inference speed of 81.4%, 74.4%, 78.1%, and 93 frames per second (FPS), which can achieve accurate and fast detection of daylily in complex field environments. The research results can provide data and experimental references for developing intelligent picking equipment for daylily.
Full article
►▼
Show Figures
Open AccessArticle
Precision Detection and Assessment of Ash Death and Decline Caused by the Emerald Ash Borer Using Drones and Deep Learning
by
Sruthi Keerthi Valicharla, Xin Li, Jennifer Greenleaf, Richard Turcotte, Christopher Hayes and Yong-Lak Park
Cited by 5 | Viewed by 1705
Abstract
Emerald ash borer (
Agrilus planipennis) is an invasive pest that has killed millions of ash trees (
Fraxinus spp.) in the USA since its first detection in 2002. Although the current methods for trapping emerald ash borers (e.g., sticky traps and
[...] Read more.
Emerald ash borer (
Agrilus planipennis) is an invasive pest that has killed millions of ash trees (
Fraxinus spp.) in the USA since its first detection in 2002. Although the current methods for trapping emerald ash borers (e.g., sticky traps and trap trees) and visual ground and aerial surveys are generally effective, they are inefficient for precisely locating and assessing the declining and dead ash trees in large or hard-to-access areas. This study was conducted to develop and evaluate a new tool for safe, efficient, and precise detection and assessment of ash decline and death caused by emerald ash borer by using aerial surveys with unmanned aerial systems (a.k.a., drones) and a deep learning model. Aerial surveys with drones were conducted to obtain 6174 aerial images including ash decline in the deciduous forests in West Virginia and Pennsylvania, USA. The ash trees in each image were manually annotated for training and validating deep learning models. The models were evaluated using the object recognition metrics: mean average precisions (
mAP) and two average precisions (
AP50 and
AP75). Our comprehensive analyses with instance segmentation models showed that Mask2former was the most effective model for detecting declining and dead ash trees with 0.789, 0.617, and 0.542 for
AP50, AP75, and
mAP, respectively, on the validation dataset. A follow-up in-situ field study conducted in nine locations with various levels of ash decline and death demonstrated that deep learning along with aerial survey using drones could be an innovative tool for rapid, safe, and efficient detection and assessment of ash decline and death in large or hard-to-access areas.
Full article
►▼
Show Figures
Open AccessArticle
Classification of Tomato Fruit Using Yolov5 and Convolutional Neural Network Models
by
Quoc-Hung Phan, Van-Tung Nguyen, Chi-Hsiang Lien, The-Phong Duong, Max Ti-Kuang Hou and Ngoc-Bich Le
Cited by 14 | Viewed by 3816
Abstract
Four deep learning frameworks consisting of Yolov5m and Yolov5m combined with ResNet50, ResNet-101, and EfficientNet-B0, respectively, are proposed for classifying tomato fruit on the vine into three categories: ripe, immature, and damaged. For a training dataset consisting of 4500 images and a training
[...] Read more.
Four deep learning frameworks consisting of Yolov5m and Yolov5m combined with ResNet50, ResNet-101, and EfficientNet-B0, respectively, are proposed for classifying tomato fruit on the vine into three categories: ripe, immature, and damaged. For a training dataset consisting of 4500 images and a training process with 200 epochs, a batch size of 128, and an image size of 224 × 224 pixels, the prediction accuracy for ripe and immature tomatoes is found to be 100% when combining Yolo5m with ResNet-101. Meanwhile, the prediction accuracy for damaged tomatoes is 94% when using Yolo5m with the Efficient-B0 model. The ResNet-50, EfficientNet-B0, Yolov5m, and ResNet-101 networks have testing accuracies of 98%, 98%, 97%, and 97%, respectively. Thus, all four frameworks have the potential for tomato fruit classification in automated tomato fruit harvesting applications in agriculture.
Full article
►▼
Show Figures
Open AccessArticle
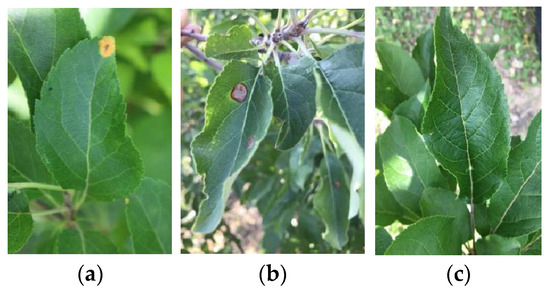
Diagnosis and Mobile Application of Apple Leaf Disease Degree Based on a Small-Sample Dataset
by
Lili Li, Bin Wang, Yanwen Li and Hua Yang
Cited by 3 | Viewed by 1808
Abstract
The accurate segmentation of apple leaf disease spots is the key to identifying the classification of apple leaf diseases and disease severity. Therefore, a DeepLabV3+ semantic segmentation network model with an actors spatial pyramid pool module (ASPP) was proposed to achieve effective extraction
[...] Read more.
The accurate segmentation of apple leaf disease spots is the key to identifying the classification of apple leaf diseases and disease severity. Therefore, a DeepLabV3+ semantic segmentation network model with an actors spatial pyramid pool module (ASPP) was proposed to achieve effective extraction of apple leaf lesion features and to improve the apple leaf disease recognition and disease severity diagnosis compared with the classical semantic segmentation network models PSPNet and GCNet. In addition, the effects of the learning rate, optimizer, and backbone network on the performance of the DeepLabV3+ network model with the best performance were analyzed. The experimental results show that the mean pixel accuracy (MPA) and mean intersection over union (MIoU) of the model reached 97.26% and 83.85%, respectively. After being deployed into the smartphone platform, the detection time of the detection system was 9s per image for the portable and intelligent diagnostics of apple leaf diseases. The transfer learning method provided the possibility of quickly acquiring a high-performance model under the condition of small datasets. The research results can provide a precise guide for the prevention and precise control of apple diseases in fields.
Full article
►▼
Show Figures
Open AccessArticle
Big Data and Machine Learning to Improve European Grapevine Moth (Lobesia botrana) Predictions
by
Joaquín Balduque-Gil, Francisco J. Lacueva-Pérez, Gorka Labata-Lezaun, Rafael del-Hoyo-Alonso, Sergio Ilarri, Eva Sánchez-Hernández, Pablo Martín-Ramos and Juan J. Barriuso-Vargas
Cited by 2 | Viewed by 2278
Abstract
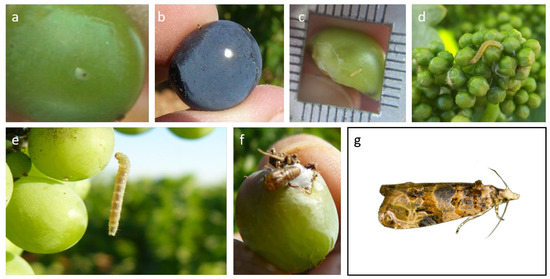
Machine Learning (ML) techniques can be used to convert Big Data into valuable information for agri-environmental applications, such as predictive pest modeling.
Lobesia botrana (Denis & Schiffermüller) 1775 (Lepidoptera: Tortricidae) is one of the main pests of grapevine, causing high productivity losses in
[...] Read more.
Machine Learning (ML) techniques can be used to convert Big Data into valuable information for agri-environmental applications, such as predictive pest modeling.
Lobesia botrana (Denis & Schiffermüller) 1775 (Lepidoptera: Tortricidae) is one of the main pests of grapevine, causing high productivity losses in some vineyards worldwide. This work focuses on the optimization of the Touzeau model, a classical correlation model between temperature and
L. botrana development using data-driven models. Data collected from field observations were combined with 30 GB of registered weather data updated every 30 min to train the ML models and make predictions on this pest’s flights, as well as to assess the accuracy of both Touzeau and ML models. The results obtained highlight a much higher F1 score of the ML models in comparison with the Touzeau model. The best-performing model was an artificial neural network of four layers, which considered several variables together and not only the temperature, taking advantage of the ability of ML models to find relationships in nonlinear systems. Despite the room for improvement of artificial intelligence-based models, the process and results presented herein highlight the benefits of ML applied to agricultural pest management strategies.
Full article
►▼
Show Figures
Open AccessArticle
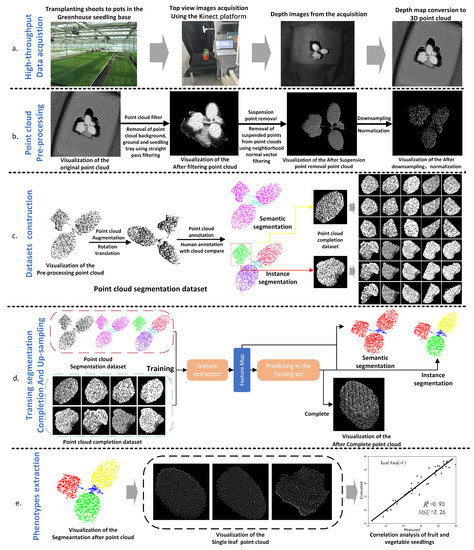
MIX-NET: Deep Learning-Based Point Cloud Processing Method for Segmentation and Occlusion Leaf Restoration of Seedlings
by
Binbin Han, Yaqin Li, Zhilong Bie, Chengli Peng, Yuan Huang and Shengyong Xu
Cited by 5 | Viewed by 2203
Abstract
In this paper, a novel point cloud segmentation and completion framework is proposed to achieve high-quality leaf area measurement of melon seedlings. In particular, the input of our algorithm is the point cloud data collected by an Azure Kinect camera from the top
[...] Read more.
In this paper, a novel point cloud segmentation and completion framework is proposed to achieve high-quality leaf area measurement of melon seedlings. In particular, the input of our algorithm is the point cloud data collected by an Azure Kinect camera from the top view of the seedlings, and our method can enhance measurement accuracy from two aspects based on the acquired data. On the one hand, we propose a neighborhood space-constrained method to effectively filter out the hover points and outlier noise of the point cloud, which can enhance the quality of the point cloud data significantly. On the other hand, by leveraging the purely linear mixer mechanism, a new network named MIX-Net is developed to achieve segmentation and completion of the point cloud simultaneously. Different from previous methods that separate these two tasks, the proposed network can better balance these two tasks in a more definite and effective way, leading to satisfactory performance on these two tasks. The experimental results prove that our methods can outperform other competitors and provide more accurate measurement results. Specifically, for the seedling segmentation task, our method can obtain a 3.1% and 1.7% performance gain compared with PointNet++ and DGCNN, respectively. Meanwhile, the
of leaf area measurement improved from 0.87 to 0.93 and
decreased from 2.64 to 2.26 after leaf shading completion.
Full article
►▼
Show Figures
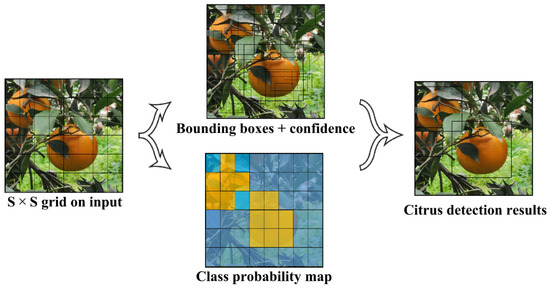
Open AccessEditor’s ChoiceArticle
A Multiscale Lightweight and Efficient Model Based on YOLOv7: Applied to Citrus Orchard
by
Junyang Chen, Hui Liu, Yating Zhang, Daike Zhang, Hongkun Ouyang and Xiaoyan Chen
Cited by 41 | Viewed by 4887
Abstract
With the gradual increase in the annual production of citrus, the efficiency of human labor has become the bottleneck limiting production. To achieve an unmanned citrus picking technology, the detection accuracy, prediction speed, and lightweight deployment of the model are important issues. Traditional
[...] Read more.
With the gradual increase in the annual production of citrus, the efficiency of human labor has become the bottleneck limiting production. To achieve an unmanned citrus picking technology, the detection accuracy, prediction speed, and lightweight deployment of the model are important issues. Traditional object detection methods often fail to achieve balanced effects in all aspects. Therefore, an improved YOLOv7 network model is proposed, which introduces a small object detection layer, lightweight convolution, and a CBAM (Convolutional Block Attention Module) attention mechanism to achieve multi-scale feature extraction and fusion and reduce the number of parameters of the model. The performance of the model was tested on the test set of citrus fruit. The average accuracy (mAP
@0.5) reached 97.29%, the average prediction time was 69.38 ms, and the number of parameters and computation costs were reduced by 11.21 M and 28.71 G compared with the original YOLOv7. At the same time, the Citrus-YOLOv7 model’s results show that it performs better compared with the current state-of-the-art network models. Therefore, the proposed Citrus-YOLOv7 model can contribute to solving the problem of citrus detection.
Full article
►▼
Show Figures
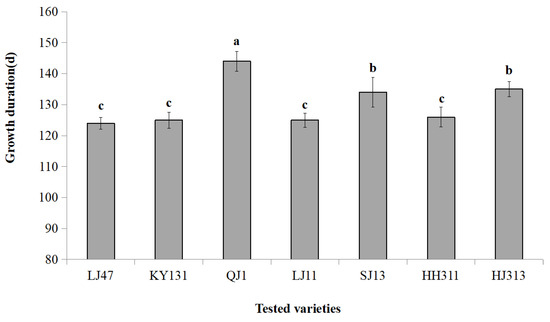
Open AccessArticle
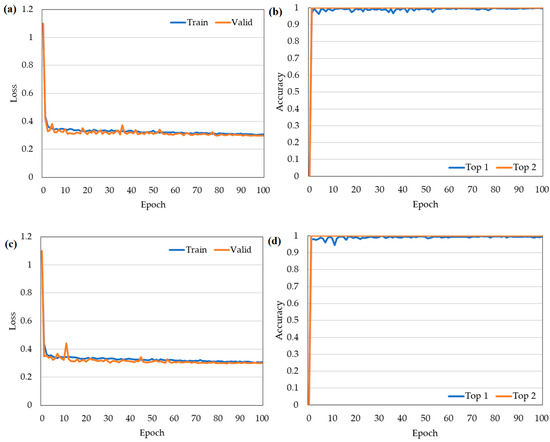
Intelligent Classification of Japonica Rice Growth Duration (GD) Based on CapsNets
by
Xin Zhao, Jianpei Zhang, Jing Yang, Bo Ma, Rui Liu and Jifang Hu
Cited by 2 | Viewed by 1456
Abstract
Rice cultivation in cold regions of China is mainly distributed in Heilongjiang Province, where the growing season of rice is susceptible to low temperature and cold damage. Choosing and planting rice varieties with suitable GD according to the accumulated temperate zone is an
[...] Read more.
Rice cultivation in cold regions of China is mainly distributed in Heilongjiang Province, where the growing season of rice is susceptible to low temperature and cold damage. Choosing and planting rice varieties with suitable GD according to the accumulated temperate zone is an important measure to prevent low temperature and cold damage. However, the traditional identification method of rice GD requires lots of field investigations, which are time consuming and susceptible to environmental interference. Therefore, an efficient, accurate, and intelligent identification method is urgently needed. In response to this problem, we took seven rice varieties suitable for three accumulated temperature zones in Heilongjiang Province as the research objects, and we carried out research on the identification of japonica rice GD based on Raman spectroscopy and capsule neural networks (CapsNets). The data preprocessing stage used a variety of methods (signal.filtfilt, difference, segmentation, and superposition) to process Raman spectral data to complete the fusion of local features and global features and data dimension transformation. A CapsNets containing three neuron layers (one convolutional layer and two capsule layers) and a dynamic routing protocol was constructed and implemented in Python. After training 160 epochs on the CapsNets, the model achieved 89% and 93% accuracy on the training and test datasets, respectively. The results showed that Raman spectroscopy combined with CapsNets can provide an efficient and accurate intelligent identification method for the classification and identification of rice GD in Heilongjiang Province.
Full article
►▼
Show Figures
Open AccessArticle
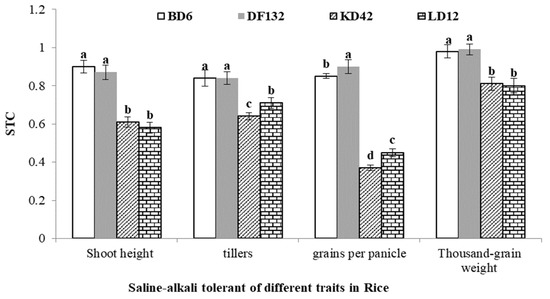
Intelligent Identification and Features Attribution of Saline–Alkali-Tolerant Rice Varieties Based on Raman Spectroscopy
by
Bo Ma, Chuanzeng Liu, Jifang Hu, Kai Liu, Fuyang Zhao, Junqiang Wang, Xin Zhao, Zhenhua Guo, Lijuan Song, Yongcai Lai and Kefei Tan
Cited by 3 | Viewed by 1673
Abstract
Planting rice in saline–alkali land can effectively improve saline–alkali soil and increase grain yield, but traditional identification methods for saline–alkali-tolerant rice varieties require tedious and time-consuming field investigations based on growth indicators by rice breeders. In this study, the Python machine deep learning
[...] Read more.
Planting rice in saline–alkali land can effectively improve saline–alkali soil and increase grain yield, but traditional identification methods for saline–alkali-tolerant rice varieties require tedious and time-consuming field investigations based on growth indicators by rice breeders. In this study, the Python machine deep learning method was used to analyze the Raman molecular spectroscopy of rice and assist in feature attribution, in order to study a fast and efficient identification method of saline–alkali-tolerant rice varieties. A total of 156 Raman spectra of four rice varieties (two saline–alkali-tolerant rice varieties and two saline–alkali-sensitive rice varieties) were analyzed, and the wave crests were extracted by an improved signal filtering difference method and the feature information of the wave crest was automatically extracted by scipy.signal.find_peaks. Select K Best (SKB), Recursive Feature Elimination (RFE) and Select F Model (SFM) were used to select useful molecular features. Based on these feature selection methods, a Logistic Regression Model (LRM) and Random Forests Model (RFM) were established for discriminant analysis. The experimental results showed that the RFM identification model based on the RFE method reached a higher recognition rate of 89.36%. According to the identification results of RFM and the identification of feature attribution materials, amylum was the most significant substance in the identification of saline–alkali-tolerant rice varieties. Therefore, an intelligent method for the identification of saline–alkali-tolerant rice varieties based on Raman molecular spectroscopy is proposed.
Full article
►▼
Show Figures
Open AccessArticle
Classification of Plant Leaves Using New Compact Convolutional Neural Network Models
by
Shivali Amit Wagle, R. Harikrishnan, Sawal Hamid Md Ali and Mohammad Faseehuddin
Cited by 18 | Viewed by 6615
Abstract
Precision crop safety relies on automated systems for detecting and classifying plants. This work proposes the detection and classification of nine species of plants of the PlantVillage dataset using the proposed developed compact convolutional neural networks and AlexNet with transfer learning. The models
[...] Read more.
Precision crop safety relies on automated systems for detecting and classifying plants. This work proposes the detection and classification of nine species of plants of the PlantVillage dataset using the proposed developed compact convolutional neural networks and AlexNet with transfer learning. The models are trained using plant leaf data with different data augmentations. The data augmentation shows a significant improvement in classification accuracy. The proposed models are also used for the classification of 32 classes of the Flavia dataset. The proposed developed N1 model has a classification accuracy of 99.45%, N2 model has a classification accuracy of 99.65%, N3 model has a classification accuracy of 99.55%, and AlexNet has a classification accuracy of 99.73% for the PlantVillage dataset. In comparison to AlexNet, the proposed models are compact and need less training time. The proposed N1 model takes 34.58%, the proposed N2 model takes 18.25%, and the N3 model takes 20.23% less training time than AlexNet. The N1 model and N3 models are size 14.8 MB making it 92.67% compact, and the N2 model is 29.7 MB which makes it 85.29% compact as compared to AlexNet. The proposed models are giving good accuracy in classifying plant leaf, as well as diseases in tomato plant leaves.
Full article
►▼
Show Figures
Open AccessReview
Bluster or Lustre: Can AI Improve Crops and Plant Health?
by
Laura-Jayne Gardiner and Ritesh Krishna
Cited by 4 | Viewed by 2614
Abstract
In a changing climate where future food security is a growing concern, researchers are exploring new methods and technologies in the effort to meet ambitious crop yield targets. The application of Artificial Intelligence (AI) including Machine Learning (ML) methods in this area has
[...] Read more.
In a changing climate where future food security is a growing concern, researchers are exploring new methods and technologies in the effort to meet ambitious crop yield targets. The application of Artificial Intelligence (AI) including Machine Learning (ML) methods in this area has been proposed as a potential mechanism to support this. This review explores current research in the area to convey the state-of-the-art as to how AI/ML have been used to advance research, gain insights, and generally enable progress in this area. We address the question—Can AI improve crops and plant health? We further discriminate the bluster from the lustre by identifying the key challenges that AI has been shown to address, balanced with the potential issues with its usage, and the key requisites for its success. Overall, we hope to raise awareness and, as a result, promote usage, of AI related approaches where they can have appropriate impact to improve practices in agricultural and plant sciences.
Full article
►▼
Show Figures
Open AccessArticle
Domain Adaptation of Synthetic Images for Wheat Head Detection
by
Zane K. J. Hartley and Andrew P. French
Cited by 12 | Viewed by 2296
Abstract
Wheat head detection is a core computer vision problem related to plant phenotyping that in recent years has seen increased interest as large-scale datasets have been made available for use in research. In deep learning problems with limited training data, synthetic data have
[...] Read more.
Wheat head detection is a core computer vision problem related to plant phenotyping that in recent years has seen increased interest as large-scale datasets have been made available for use in research. In deep learning problems with limited training data, synthetic data have been shown to improve performance by increasing the number of training examples available but have had limited effectiveness due to
domain shift. To overcome this, many adversarial approaches such as Generative Adversarial Networks (GANs) have been proposed as a solution by better aligning the distribution of synthetic data to that of real images through domain augmentation. In this paper, we examine the impacts of performing wheat head detection on the global wheat head challenge dataset using synthetic data to supplement the original dataset. Through our experimentation, we demonstrate the challenges of performing domain augmentation where the target domain is large and diverse. We then present a novel approach to improving scores through using heatmap regression as a support network, and clustering to combat high variation of the target domain.
Full article
►▼
Show Figures
Planned Papers
The below list represents only planned manuscripts. Some of these
manuscripts have not been received by the Editorial Office yet. Papers
submitted to MDPI journals are subject to peer-review.
Title: Bluster or lustre: Can AI improve crops and plant health?
Authors: Laura-Jayne Gardiner; Ritesh Krishna
Affiliation: IBM Research, The Hartree Centre, Warrington, WA4 4AD, UK
Abstract: In a changing climate where future food security is a growing concern, researchers are exploring new methods and technologies in the effort to meet ambitious crop yield targets. The application of Artificial Intelligence (AI) including Machine Learning (ML) methods in this area has been proposed as a potential mechanism to support this. This review explores current research in the area to convey the state-of-the-art as to how AI/ML have been used to advance research, gain insights and generally enable progress in this area. We address the question - Can AI improve crops and plant health? We further discriminate the bluster from the lustre by identifying the key challenges that AI has been shown to address, balanced with the potential issues with its usage, and the key requisites for its success. Overall, we hope to raise awareness and, as a result, promote usage, of AI related approaches where they can have appropriate impact to improve practices in agricultural and plant sciences.