Bovine Leukemia Virus Infection
A topical collection in Pathogens (ISSN 2076-0817). This collection belongs to the section "Viral Pathogens".
Viewed by 47817
Share This Topical Collection
Editors
 Dr. Tasia M. (Taxis) Kendrick
Dr. Tasia M. (Taxis) Kendrick
 Dr. Tasia M. (Taxis) Kendrick
Dr. Tasia M. (Taxis) Kendrick
E-Mail
Website
Collection Editor
Department of Animal Science, College of Agriculture and Natural Resources, Michigan State University, East Lansing, MI 48824, USA
Interests: genetics/genomics; bovine leukemia virus; dairy science; education research
 Prof. Dr. Paul C. Bartlett
Prof. Dr. Paul C. Bartlett
 Prof. Dr. Paul C. Bartlett
Prof. Dr. Paul C. Bartlett
E-Mail
Website
Collection Editor
College of Veterinary Medicine, Michigan State University, East Lansing, MI 48824, USA
Interests: controlling bovine leukemia virus in cattle populations
Topical Collection Information
Dear Colleagues,
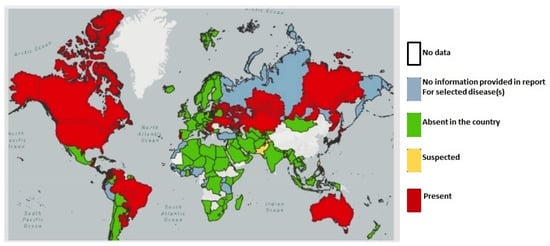
Enzootic bovine leukosis caused by the bovine leukemia virus (BLV) has been eradicated in over 20 countries, most of which are in Western Europe. In the last several years there have been major changes in our understanding of BLV epidemiology and major advancements in diagnosis and disease control methods. The purpose of this Topical Collection is to highlight these recent developments. Surveys outside of Europe have shown dramatic increases in BLV prevalence in both beef and dairy cattle. The subclinical impact on milk production, cow lifespan (longevity) and immune function has only recently been documented. Infected cattle appear predisposed to mastitis and lameness, and animal welfare is clearly negatively affected. BLV DNA has reportedly been associated with human mammary cancer, re-opening public health concerns which were once considered resolved many decades ago. All these developments have spurred an increased interest in controlling BLV in cattle. Many nations now have 30-50% of their cattle infected, so it may be economically impossible for them to follow the traditional method of eradicating BLV by culling all antibody-positive cattle. Alternative control approaches are being sought such as vaccination, host genetic selection and prevalence reduction through management interventions. Recent development of qPCR tests to measure BLV proviral load are showing some application in reducing transmission by identifying the most infectious cattle for segregation or culling. Researchers actively engaged in studying BLV epidemiology, immunology, molecular biology, virology, pathology, etc. are enthusiastically encouraged to submit their works to this Topical Collection.
Dr. Tasia M. (Taxis) Kendrick
Prof. Dr. Paul C. Bartlett
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Pathogens is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Published Papers (20 papers)
Open AccessArticle
Genetic Variability of Bovine Leukemia Virus: Evidence of Dual Infection, Recombination and Quasi-Species
by
Aneta Pluta, Marzena Rola-Łuszczak, Federico G. Hoffmann, Irina Donnik, Maxim Petropavlovskiy and Jacek Kuźmak
Viewed by 998
Abstract
We have characterized the intrahost genetic variation in the bovine leukemia virus (BLV) by examining 16 BLV isolates originating from the Western Siberia–Tyumen and South Ural–Chelyabinsk regions of Russia. Our research focused on determining the genetic composition of an 804 bp fragment of
[...] Read more.
We have characterized the intrahost genetic variation in the bovine leukemia virus (BLV) by examining 16 BLV isolates originating from the Western Siberia–Tyumen and South Ural–Chelyabinsk regions of Russia. Our research focused on determining the genetic composition of an 804 bp fragment of the BLV
env gene, encoding for the entire gp51 protein. The results provide the first indication of the quasi-species genetic nature of BLV infection and its relevance for genome-level variation. Furthermore, this is the first phylogenetic evidence for the existence of a dual infection with BLV strains belonging to different genotypes within the same host: G4 and G7. We identified eight cases of recombination between these two BLV genotypes. The detection of quasi-species with cases of dual infection and recombination indicated a higher potential of BLV for genetic variability at the intra-host level than was previously considered.
Full article
►▼
Show Figures
Open AccessArticle
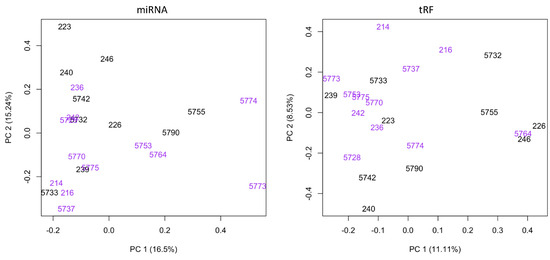
Influence of Maternal BLV Infection on miRNA and tRF Expression in Calves
by
Anna K. Goldkamp, Ciarra H. Lahuis, Darren E. Hagen and Tasia M. Taxis
Viewed by 973
Abstract
Small non-coding RNAs, such as microRNAs (miRNA) and tRNA-derived fragments (tRF), are known to be involved in post-transcriptional gene regulation. Research has provided evidence that small RNAs may influence immune development in calves. Bovine leukosis is a disease in cattle caused by Bovine
[...] Read more.
Small non-coding RNAs, such as microRNAs (miRNA) and tRNA-derived fragments (tRF), are known to be involved in post-transcriptional gene regulation. Research has provided evidence that small RNAs may influence immune development in calves. Bovine leukosis is a disease in cattle caused by Bovine Leukemia Virus (BLV) that leads to increased susceptibility to opportunistic pathogens. No research has addressed the potential influence that a maternal BLV infection may have on gene regulation through the differential expression of miRNAs or tRFs in progeny. Blood samples from 14-day old Holstein calves born to BLV-infected dams were collected. Antibodies for BLV were assessed using ELISA and levels of BLV provirus were assessed using qPCR. Total RNA was extracted from whole blood samples for small RNA sequencing. Five miRNAs (bta-miR-1, bta-miR-206, bta-miR-133a, bta-miR-133b, and bta-miR-2450d) and five tRFs (tRF-36-8JZ8RN58X2NF79E, tRF-20-0PF05B2I, tRF-27-W4R951KHZKK, tRF-22-S3M8309NF, and tRF-26-M87SFR2W9J0) were dysregulated in calves born to BLV-infected dams. The miRNAs appear to be involved in the gene regulation of immunological responses and muscle development. The tRF subtypes and parental tRNA profiles in calves born to infected dams appear to be consistent with previous publications in adult cattle with BLV infection. These findings offer insight into how maternal BLV infection status may impact the development of offspring.
Full article
►▼
Show Figures
Open AccessArticle
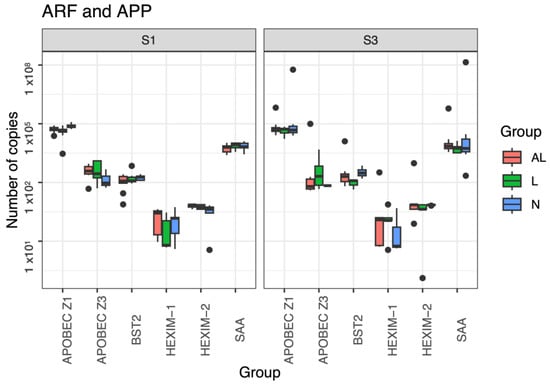
Study of the Genetic Expression of Antiretroviral Restriction Factors and Acute Phase Proteins in Cattle Infected with Bovine Leukemia Virus
by
Ana S. González-Méndez, Jorge L. Tórtora Pérez, Edith Rojas-Anaya and Hugo Ramírez Álvarez
Viewed by 1272
Abstract
The goal of this study was to analyze the genetic expression of antiretroviral restriction factors (ARF) and acute phase proteins (APP), as well as their correlation with proviral and viral loads in cattle with aleukemic (AL) and persistent lymphocytosis (PL). Complete blood samples
[...] Read more.
The goal of this study was to analyze the genetic expression of antiretroviral restriction factors (ARF) and acute phase proteins (APP), as well as their correlation with proviral and viral loads in cattle with aleukemic (AL) and persistent lymphocytosis (PL). Complete blood samples were collected from a herd of dairy cows, and we extracted genetic material from peripheral blood leukocytes. Absolute quantification of the expression of ARF (APOBEC-Z1, Z2, and Z3; HEXIM-1, HEXIM-2, and BST2) and APP (haptoglobin (HP), and serum amyloid A (SAA)) was performed by qPCR. Statistical significance was observed in the expression of APOBEC-Z3 in BLV-infected animals. We only found positive correlations with a strong expression of the ARF genes in the AL group. The participation of APOBEC (Z1 and Z3), HEXIM-1, and HEXIM-2 was more frequently identified in BLV-infected animals. HEXIM-2 showed active gene expression in the AL group. Although the expression of ARF in early stages of infection (AL) maintains an important participation, in late stages (PL) it seems to have little relevance.
Full article
►▼
Show Figures
Open AccessArticle
Identification of BoLA Alleles Associated with BLV Proviral Load in US Beef Cows
by
Ciarra H. LaHuis, Oscar J. Benitez, Casey J. Droscha, Sukhdeep Singh, Andrew Borgman, Chaelynne E. Lohr, Paul C. Bartlett and Tasia M. Taxis
Cited by 3 | Viewed by 1543
Abstract
Bovine leukemia virus (BLV) causes enzootic bovine leukosis, the most common neoplastic disease in cattle. Previous work estimates that 78% of US beef operations and 38% of US beef cattle are seropositive for BLV. Infection by BLV in a herd is an economic
[...] Read more.
Bovine leukemia virus (BLV) causes enzootic bovine leukosis, the most common neoplastic disease in cattle. Previous work estimates that 78% of US beef operations and 38% of US beef cattle are seropositive for BLV. Infection by BLV in a herd is an economic concern for producers as evidence suggests that it causes an increase in cost and a subsequent decrease in profit to producers. Studies investigating BLV in dairy cattle have noted disease resistance or susceptibility, measured by a proviral load (PVL) associated with specific alleles of the bovine leukocyte antigen (BoLA) DRB3 gene. This study aims to investigate the associations between BoLA DRB3 alleles and BLV PVL in beef cattle. Samples were collected from 157 Midwest beef cows. BoLA DRB3 alleles were identified and compared with BLV PVL. One BoLA DRB3 allele,
*026:01, was found to be associated with high PVL in relation to the average of the sampled population. In contrast, two alleles,
*033:01 and
*002:01, were found to be associated with low PVL. This study provides evidence of a relationship between BoLA DRB3 alleles and BLV PVL in US beef cows.
Full article
►▼
Show Figures
Open AccessArticle
BoLA-DRB3 Polymorphism Controls Proviral Load and Infectivity of Bovine Leukemia Virus (BLV) in Milk
by
Ayumi Nakatsuchi, Sonoko Watanuki, Liushiqi Borjigin, Hirotaka Sato, Lanlan Bai, Ryosuke Matsuura, Maho Kuroda, Hironobu Murakami, Reiichiro Sato, Sakurako Asaji, Asako Ando, Yasunobu Matsumoto, Shin-Nosuke Takeshima and Yoko Aida
Cited by 12 | Viewed by 1817
Abstract
Bovine leukemia virus (BLV), which causes enzootic bovine leukosis, is transmitted to calves through the milk of BLV-infected dams. Bovine leukocyte antigen (BoLA)-
DRB3 is a polymorphic gene associated with BLV infectivity and proviral load (PVL). However, the effect of
BoLA-DRB3 polymorphism on
[...] Read more.
Bovine leukemia virus (BLV), which causes enzootic bovine leukosis, is transmitted to calves through the milk of BLV-infected dams. Bovine leukocyte antigen (BoLA)-
DRB3 is a polymorphic gene associated with BLV infectivity and proviral load (PVL). However, the effect of
BoLA-DRB3 polymorphism on the infectivity and PVL of milk from BLV-infected dams remains unknown. This study examined milk from 259 BLV-infected dams, including susceptible dams carrying at least one
BoLA-DRB3*012:01 or
*015:01 allele with high PVL, resistant dams carrying at least one
BoLA-DRB3*002:01,
*009:02, or
*014:01:01 allele with low PVL, and neutral dams carrying other alleles. The detection rate of BLV provirus and PVL were significantly higher in milk from susceptible dams than in that from resistant dams. This result was confirmed in a three-year follow-up study in which milk from susceptible dams showed a higher BLV provirus detection rate over a longer period than that from resistant dams. The visualization of infectivity of milk cells using a luminescence syncytium induction assay showed that the infectious risk of milk from BLV-infected dams was markedly high for susceptible dams compared to resistant ones. This is the first report confirming that
BoLA-DRB3 polymorphism affects the PVL and infectivity of milk from BLV-infected dams.
Full article
►▼
Show Figures
Open AccessArticle
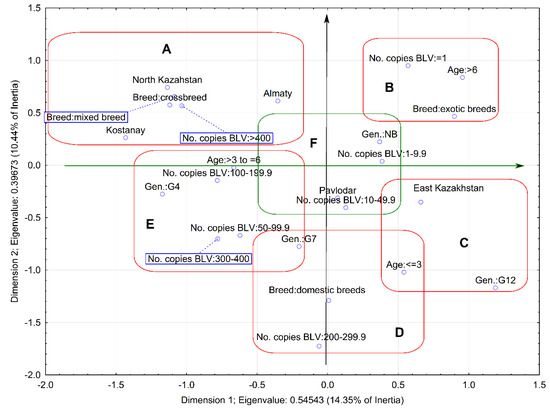
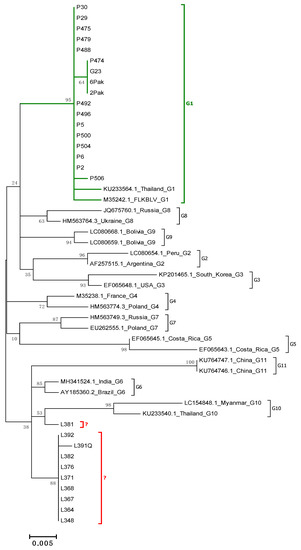
Molecular Characterization of Bovine Leukemia Virus with the Evidence of a New Genotype Circulating in Cattle from Kazakhstan
by
Akhmetzhan Sultanov, Marzena Rola-Łuszczak, Saltanat Mamanova, Anna Ryło, Zbigniew Osiński, Meruyert A. Saduakassova, Elvira Bashenova and Jacek Kuźmak
Cited by 11 | Viewed by 2615
Abstract
Bovine leukemia virus (BLV) is a retrovirus that causes enzootic bovine leukosis (EBL) and has worldwide distribution. Infections with BLV have been reported in cattle from Kazakhstan but the virus has not yet been thoroughly characterized. In this study, we detect and estimate
[...] Read more.
Bovine leukemia virus (BLV) is a retrovirus that causes enzootic bovine leukosis (EBL) and has worldwide distribution. Infections with BLV have been reported in cattle from Kazakhstan but the virus has not yet been thoroughly characterized. In this study, we detect and estimate the level of BLV proviral DNA by qPCR in DNA samples from 119 cattle naturally infected with BLV, from 18 farms located in four different geographical regions of Kazakhstan. Furthermore, we conducted the phylogenetic and molecular analysis of 41 BLV env-gp51 gene sequences from BLV infected cattle. Phylogenetic analysis showed the affiliation of sequences to two already known genotypes G4 and G7 and also to a new genotype, classified as genotype G12. In addition, a multivariate method was employed for analysis of the association between proviral load and different variables such as the geographical location of the herd, cattle breeds, age of animals, and the presence of particular BLV genotypes. In summary, the results of this study provide the first evidence on molecular characterization of BLV circulating in cattle from Kazakhstan.
Full article
►▼
Show Figures
Open AccessArticle
Phenotypic Selection of Dairy Cattle Infected with Bovine Leukemia Virus Demonstrates Immunogenetic Resilience through NGS-Based Genotyping of BoLA MHC Class II Genes
by
Chaelynne E. Lohr, Kelly R. B. Sporer, Kelsey A. Brigham, Laura A. Pavliscak, Matelyn M. Mason, Andrew Borgman, Vickie J. Ruggiero, Tasia M. Taxis, Paul C. Bartlett and Casey J. Droscha
Cited by 4 | Viewed by 2949
Abstract
Characterization of the bovine leukocyte antigen (BoLA) DRB3 gene has shown that specific alleles associate with susceptibility or resilience to the progression of bovine leukemia virus (BLV), measured by proviral load (PVL). Through surveillance of multi-farm BLV eradication field trials, we observed differential
[...] Read more.
Characterization of the bovine leukocyte antigen (BoLA) DRB3 gene has shown that specific alleles associate with susceptibility or resilience to the progression of bovine leukemia virus (BLV), measured by proviral load (PVL). Through surveillance of multi-farm BLV eradication field trials, we observed differential phenotypes within seropositive cows that persist from months to years. We sought to develop a multiplex next-generation sequencing workflow (NGS-SBT) capable of genotyping 384 samples per run to assess the relationship between BLV phenotype and two BoLA genes. We utilized longitudinal results from milk ELISA screening and subsequent blood collections on seropositive cows for PVL determination using a novel BLV proviral load multiplex qPCR assay to phenotype the cows. Repeated diagnostic observations defined two distinct phenotypes in our study population, ELISA-positive cows that do not harbor detectable levels of provirus and those who do have persistent proviral loads. In total, 565 cows from nine Midwest dairy farms were selected for NGS-SBT, with 558 cows: 168 BLV susceptible (ELISA-positive/PVL-positive) and 390 BLV resilient (ELISA-positive/PVL-negative) successfully genotyped. Three BoLA-DRB3 alleles, including one novel allele, were shown to associate with disease resilience,
*009:02,
*044:01, and
*048:02 were found at rates of 97.5%, 86.5%, and 90.3%, respectively, within the phenotypically resilient population. Alternatively,
DRB3*015:01 and
*027:03, both known to associate with disease progression, were found at rates of 81.1% and 92.3%, respectively, within the susceptible population. This study helps solidify the immunogenetic relationship between BoLA-DRB3 alleles and BLV infection status of these two phenotypic groupings of US dairy cattle.
Full article
►▼
Show Figures
Open AccessArticle
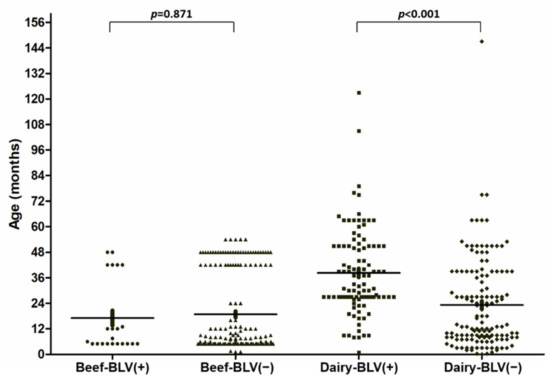
Potential Risk Factors Associated with Infection with Bovine Leukaemia Virus in Dairy and Beef Cattle in Taiwan
by
Yi-Chen Chen, Wen-Yu Chin, Chao-Chin Chang, Shih-Te Chuang and Wei-Li Hsu
Cited by 2 | Viewed by 1716
Abstract
Bovine leukaemia virus (BLV), which is classified as a
Deltaretrovirus, is the aetiologic agent of enzootic bovine leukosis (EBL), a chronic lymphoproliferative disorder with a worldwide distribution. EBL is widespread in dairy herds and causes a direct economic impact due to reduced
[...] Read more.
Bovine leukaemia virus (BLV), which is classified as a
Deltaretrovirus, is the aetiologic agent of enzootic bovine leukosis (EBL), a chronic lymphoproliferative disorder with a worldwide distribution. EBL is widespread in dairy herds and causes a direct economic impact due to reduced milk production and the early culling of BLV-infected cattle. The BLV infection status in Taiwan remains largely unknown; a high prevalence of BLV in dairy cows was recently revealed. The present study further investigated BLV infections in beef cattle. Surprisingly, the prevalence of BLV proviral DNA was as low as 11.8% (23/195), which is significantly lower than that noted in dairy cows, which was 42.5% (102/240) (
p < 0.001). Factors associated with BLV infections were subsequently investigated. Due to the differences in herd management, an analysis of risk factors for a BLV infection was independently conducted in these two sectors. Several factors associated with a BLV infection were identified. Age was significantly associated with BLV infection status in dairy cows (
p < 0.001) but not in beef cattle. A high prevalence of BLV was observed in cattle >15.5 months old (57.8%) compared with those ≤15.5 months old (11.4%). Moreover, after stratification analysis, based on the critical age of 15.5 months, as determined by the receiver operating characteristic (ROC) curve, a significantly higher BLV prevalence was demonstrated in lactating dairy cows, cattle undergoing bull breeding, heifers at older ages, and those undergoing routine rectal palpation. Due to the high prevalence of BLV in Taiwan, the development of an effective control program, based on the identified risk factors, is important for interrupting the routes of BLV transmission within herds.
Full article
►▼
Show Figures
Open AccessArticle
Enzootic Bovine Leukosis in Italy: Epidemiological Issues after Free Status Recognition and Measures Applied to Tackle the Last Persistent Clusters
by
Cecilia Righi, Carmen Iscaro, Stefano Petrini, Roberto Lomolino and Francesco Feliziani
Cited by 1 | Viewed by 1839
Abstract
Enzootic Bovine Leukosis (EBL), caused by the bovine leukemia virus (BLV), has been eradicated in over 20 countries, most of which are in Western Europe. The European Commission, in 2017, declared Italy to be an officially EBL-free country by means of Commission Implementing
[...] Read more.
Enzootic Bovine Leukosis (EBL), caused by the bovine leukemia virus (BLV), has been eradicated in over 20 countries, most of which are in Western Europe. The European Commission, in 2017, declared Italy to be an officially EBL-free country by means of Commission Implementing Decision (EU) 2017/1910, despite the presence of some infection clusters located in four regions of Central-Southern Italy. As a consequence of persisting infection, the Italian Ministry of Health established specific eradication measures in these areas. In collaboration with the National Reference Laboratory for the Study of Ruminant Retroviral Infectious Diseases, the Ministry of Health employed data from the veterinary information system digital platform, combined with a gap analysis exercise, to monitor and verify the progress of control activities within infection clusters during the period 2018–2021. Our aim was to identify any remaining gaps and, consequently, specific measures to eliminate the factors favouring EBL persistence, on the basis of a description and analysis of the current data regarding epidemiological trends in Italian clusters. The final goal is to achieve the implementation of a less expensive surveillance plan in these areas, as well. The results of comprehensive analysis showed that the eradication activities had been effectively implemented by official local veterinary services, resulting in a drastic reduction of EBL outbreaks in most territories during the period 2018–2021.
Full article
►▼
Show Figures
Open AccessArticle
Kinetic Study of BLV Infectivity in BLV Susceptible and Resistant Cattle in Japan from 2017 to 2019
by
Lanlan Bai, Liushiqi Borjigin, Hirotaka Sato, Shin-Nosuke Takeshima, Sakurako Asaji, Hiroshi Ishizaki, Keiji Kawashima, Yuko Obuchi, Shinji Sunaga, Asako Ando, Hidehito Inoko, Satoshi Wada and Yoko Aida
Cited by 12 | Viewed by 1931
Abstract
Bovine leukemia virus (BLV) is the causative agent of enzootic bovine leukosis. Polymorphism in bovine lymphocyte antigen
(BoLA)-DRB3 alleles is related to susceptibility to BLV proviral load (PVL), which is a useful index for estimating disease progression and transmission risk. However, whether differential
[...] Read more.
Bovine leukemia virus (BLV) is the causative agent of enzootic bovine leukosis. Polymorphism in bovine lymphocyte antigen
(BoLA)-DRB3 alleles is related to susceptibility to BLV proviral load (PVL), which is a useful index for estimating disease progression and transmission risk. However, whether differential
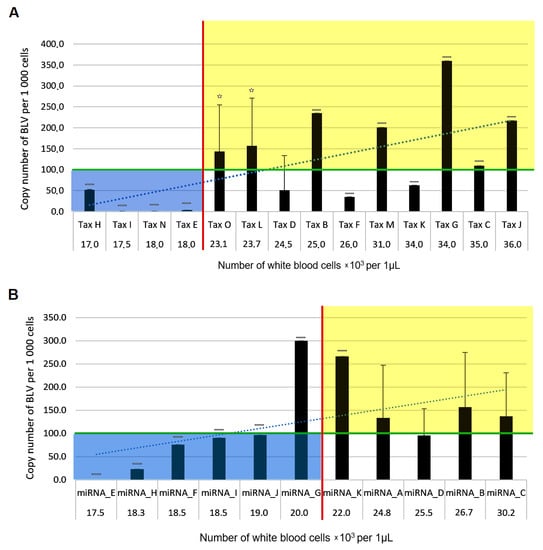
BoLA-DRB3 affects BLV infectivity remains unknown. In a three-year follow-up investigation using a luminescence syncytium induction assay for evaluating BLV infectivity, we visualized and evaluated the kinetics of BLV infectivity in cattle with susceptible, resistant and neutral
BoLA-DRB3 alleles which were selected from 179 cattle. Susceptible cattle showed stronger BLV infectivity than both resistant and neutral cattle. The order of intensity of BLV infectivity was as follows: susceptible cattle > neutral cattle > resistant cattle. BLV infectivity showed strong positive correlation with PVL at each testing point. BLV-infected susceptible cattle were found to be at higher risk of horizontal transmission, as they had strong infectivity and high PVL, whereas BLV-infected resistant cattle were low risk of BLV transmission owing to weak BLV infection and low PVL. Thus, this is the first study to demonstrate that the
BoLA-DRB3 polymorphism is associated with BLV infection.
Full article
►▼
Show Figures
Open AccessArticle
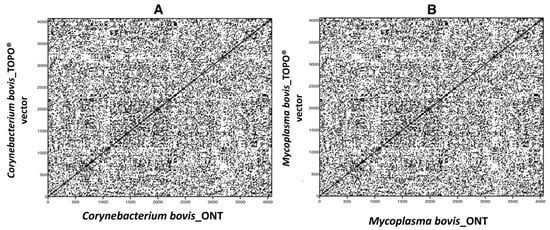
Tracing Viral Transmission and Evolution of Bovine Leukemia Virus through Long Read Oxford Nanopore Sequencing of the Proviral Genome
by
Laura A. Pavliscak, Jayaveeramuthu Nirmala, Vikash K. Singh, Kelly R. B. Sporer, Tasia M. Taxis, Pawan Kumar, Sagar M. Goyal, Sunil Kumar Mor, Declan C. Schroeder, Scott J. Wells and Casey J. Droscha
Cited by 5 | Viewed by 3217
Abstract
Bovine leukemia virus (BLV) causes Enzootic Bovine Leukosis (EBL), a persistent life-long disease resulting in immune dysfunction and shortened lifespan in infected cattle, severely impacting the profitability of the US dairy industry. Our group has found that 94% of dairy farms in the
[...] Read more.
Bovine leukemia virus (BLV) causes Enzootic Bovine Leukosis (EBL), a persistent life-long disease resulting in immune dysfunction and shortened lifespan in infected cattle, severely impacting the profitability of the US dairy industry. Our group has found that 94% of dairy farms in the United States are infected with BLV with an average in-herd prevalence of 46%. This is partly due to the lack of clinical presentation during the early stages of primary infection and the elusive nature of BLV transmission. This study sought to validate a near-complete genomic sequencing approach for reliability and accuracy before determining its efficacy in characterizing the sequence identity of BLV proviral genomes collected from a pilot study made up of 14 animals from one commercial dairy herd. These BLV-infected animals were comprised of seven adult dam/daughter pairs that tested positive by ELISA and qPCR. The results demonstrate sequence identity or divergence of the BLV genome from the same samples tested in two independent laboratories, suggesting both vertical and horizontal transmission in this dairy herd. This study supports the use of Oxford Nanopore sequencing for the identification of viral SNPs that can be used for retrospective genetic contact tracing of BLV transmission.
Full article
►▼
Show Figures
Open AccessArticle
Diagnostic Measures of Disease Progression in Cattle Following Natural Infection with Bovine Leukemia Virus
by
Holden C. Hutchinson, Vickie J. Ruggiero, Bo Norby, Kelly R. B. Sporer and Paul C. Bartlett
Cited by 3 | Viewed by 1742
Abstract
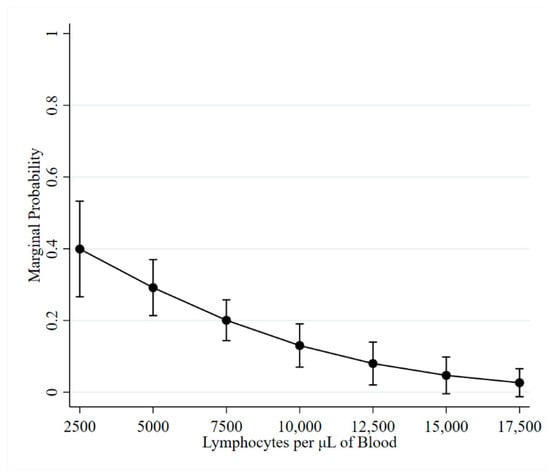
This study describes the longitudinal changes in bovine leukemia virus (BLV) ELISA antibodies, proviral load (PVL), and blood lymphocyte counts (LC) observed over a 2.5-year period in naturally infected cattle. The dataset utilized was from a BLV intervention field trial on three Midwestern
[...] Read more.
This study describes the longitudinal changes in bovine leukemia virus (BLV) ELISA antibodies, proviral load (PVL), and blood lymphocyte counts (LC) observed over a 2.5-year period in naturally infected cattle. The dataset utilized was from a BLV intervention field trial on three Midwestern dairy herds. Our analysis showed ELISA false negatives were more likely to occur in cattle with low PVL and normal LC. On average, negligible changes in LC were observed during six-month intervals. Periods of lymphocytosis, defined as >10,000 lymphocytes per uL of blood, were observed in 31.5% (68/216) of BLV test-positive cattle. In BLV test-positive cows, an average increase of 2900 to 3100 proviral copies per 100,000 cells was observed during each subsequent six-month sampling interval. The difference between the minimum and maximum PVL observed for an ELISA-positive cow with 3 or more observations ranged from 0 to 115,600 copies per 100,000 cells (median: 12,900; mean: 19,200). Therefore, following the identification of ELISA-positive cattle and the assessment of PVL and LC, subsequent semiannual tests to assess disease progression may not be needed. Further work is needed to determine how available diagnostic tests can be optimized to design cost-effective testing schemes for BLV control programs.
Full article
►▼
Show Figures
Open AccessArticle
Molecular Characterization of the env Gene of Bovine Leukemia Virus in Cattle from Pakistan with NGS-Based Evidence of Virus Heterogeneity
by
Marzena Rola-Łuszczak, Ali Sakhawat, Aneta Pluta, Anna Ryło, Arkadiusz Bomba, Nazia Bibi and Jacek Kuźmak
Cited by 6 | Viewed by 2916
Abstract
Characterization of the global genetic diversity of the bovine leukemia virus (BLV) is an ongoing international research effort. Up to now BLV sequences have been classified into eleven distinct genotypes. Although BLV genotyping and molecular analysis of field isolates were reported in many
[...] Read more.
Characterization of the global genetic diversity of the bovine leukemia virus (BLV) is an ongoing international research effort. Up to now BLV sequences have been classified into eleven distinct genotypes. Although BLV genotyping and molecular analysis of field isolates were reported in many countries, there is no report describing BLV genotypes present in cattle from Pakistan. In this study we examined 27
env gene sequences from BLV-infected cattle coming from four farms located in Khyber Pakhtunkwa, Gilgit Baltisan and Punjab provinces. Phylogenetic analyses revealed the classification of Pakistani sequences into genotypes G1 and G6. The alignment with the FLK-BLV sequence revealed the presence of 45 mutations, namely, seven in genotype G1 and 33 in genotype G6. Five mutations were found in both, G1 and G6 genotypes. Twelve amino acid substitutions were found in the analyzed sequences, of which only one P264S was specific for sequences from Pakistan. Furthermore, a certain degree of nucleotide heterogeneity was identified by NGS. These results highlight the need for further study on the importance of genetic variability of BLV, especially in the context of its pathogenicity and potential effect on serological detection.
Full article
►▼
Show Figures
Open AccessArticle
Natural Infection of Dairy Cows with Bovine Leukemia Virus Affects Immunoglobulin Levels in Saliva and Serum but Not Milk
by
Monika Dziuba, Vickie J. Ruggiero, Catherine Wilson, Paul C. Bartlett and Paul M. Coussens
Cited by 1 | Viewed by 2192
Abstract
Bovine leukemia virus (BLV) is a retroviral infection that disrupts the immune function of infected animals. It is widespread among U.S. dairy cattle. In this pilot study, the average total IgA and IgM concentrations in milk, saliva, and serum samples from BLV ELISA-positive
[...] Read more.
Bovine leukemia virus (BLV) is a retroviral infection that disrupts the immune function of infected animals. It is widespread among U.S. dairy cattle. In this pilot study, the average total IgA and IgM concentrations in milk, saliva, and serum samples from BLV ELISA-positive (ELISA+) dairy cows were compared against samples from BLV ELISA-negative (ELISA−) cows using the Kruskal–Wallis test (with ties). The results from ELISA+ cows were also stratified by lymphocyte count (LC) and proviral load (PVL). In milk and saliva from ELISA+ cows, the average total IgA and IgM concentrations were decreased compared to ELISA− cows, although this was only statistically significant for saliva IgM in cows with low PVL (
p = 0.0424). Numerically, the average total IgA concentrations were 33.6% lower in milk and 23.7% lower in saliva, and the average total IgM concentrations were 42.4% lower in milk and 15.5% lower in saliva. No significant differences were observed in the total serum IgA concentrations, regardless of PVL and LC. The total serum IgM from ELISA+ cows was significantly decreased (
p = 0.0223), with the largest decreases occurring in the highest PVL and LC subgroups. This pilot study is a first step in investigating the impact of BLV on mucosal immunity and will require further exploration in each of the various stages of disease progression.
Full article
Open AccessArticle
Risk Assessment of Bovine Major Histocompatibility Complex Class II DRB3 Alleles for Perinatal Transmission of Bovine Leukemia Virus
by
Liushiqi Borjigin, Chieh-Wen Lo, Lanlan Bai, Rania Hamada, Hirotaka Sato, Shuji Yoneyama, Anna Yasui, Sohei Yasuda, Risa Yamanaka, Munehito Mimura, Michihito Inokuma, Yasuo Shinozaki, Naoko Tanaka, Shin-Nosuke Takeshima and Yoko Aida
Cited by 14 | Viewed by 1846
Abstract
Perinatal transmission plays a critical role in the spread of bovine leukemia virus (BLV) infection in cattle herds. In the Holstein breed, we previously identified BLV resistant and susceptible bovine leukocyte antigen (
BoLA)-
DRB3 alleles, including
BoLA-
DRB3*009:02 and
*014:01:01
[...] Read more.
Perinatal transmission plays a critical role in the spread of bovine leukemia virus (BLV) infection in cattle herds. In the Holstein breed, we previously identified BLV resistant and susceptible bovine leukocyte antigen (
BoLA)-
DRB3 alleles, including
BoLA-
DRB3*009:02 and
*014:01:01 with a low BLV proviral load (PVL), and
*015:01 and
*012:01 with a high PVL. Here, we evaluated the perinatal BLV transmission risk in dams with different
BoLA-DRB3 alleles.
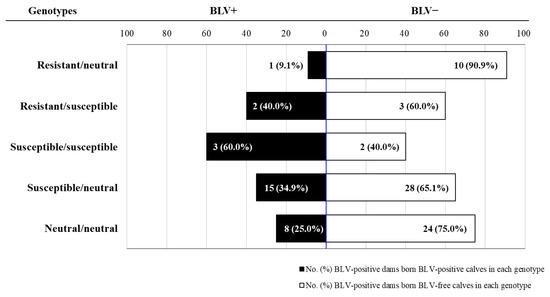
BoLA-DRB3 alleles of 120 dam-calf pairs from five dairy farms in Japan were identified; their PVL was quantified using the BLV-Coordination of Common Motifs (CoCoMo)-qPCR-2 assay. Ninety-six dams were BLV-positive, and 29 gave birth to BLV-infected calves. Perinatal transmission frequency was 19% in dams with resistant alleles suppressed to a low PVL level, and 38% and 25% in dams with susceptible and neutral alleles that maintained high PVL levels, respectively. Notably, all calves with resistant alleles were BLV free, whereas 30% of calves with susceptible genes were infected. Thus, vertical transmission risk was extremely lower for dams and calves with resistant alleles compared to those with susceptible alleles. Our results can inform the development of effective BLV eradication programs under field conditions by providing necessary data to allow for optimal selection of dams for breeding.
Full article
►▼
Show Figures
Open AccessArticle
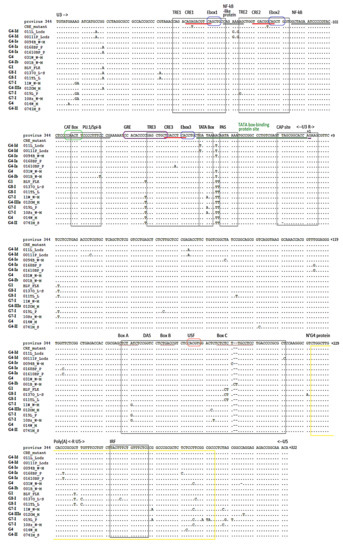
Analysis of Nucleotide Sequence of Tax, miRNA and LTR of Bovine Leukemia Virus in Cattle with Different Levels of Persistent Lymphocytosis in Russia
by
Aneta Pluta, Natalia V. Blazhko, Charity Ngirande, Thomas Joris, Luc Willems and Jacek Kuźmak
Cited by 8 | Viewed by 2831
Abstract
Bovine Leukemia Virus (BLV) is the etiological agent of enzootic bovine leucosis (EBL), a lymphoproliferative disease of the bovine species. In BLV-infected cells, the long terminal repeat (LTR), the viral Tax protein and viral miRNAs promote viral and cell proliferation as well as
[...] Read more.
Bovine Leukemia Virus (BLV) is the etiological agent of enzootic bovine leucosis (EBL), a lymphoproliferative disease of the bovine species. In BLV-infected cells, the long terminal repeat (LTR), the viral Tax protein and viral miRNAs promote viral and cell proliferation as well as tumorigenesis. Although their respective roles are decisive in BLV biology, little is known about the genetic sequence variation of these parts of the BLV genome and their impact on disease outcome. Therefore, the objective of this study was to assess the relationship between disease progression and sequence variation of the BLV Tax, miRNA and LTR regions in infected animals displaying either low or high levels of persistent lymphocytosis (PL). A statistically significant association was observed between the A(+187)C polymorphism in the downstream activator sequence (DAS) region in LTR (
p-value = 0.00737) and high lymphocytosis. Our study also showed that the mutation A(−4)G in the CAP site occurred in 70% of isolates with low PL and was not found in the high PL group. Conversely, the mutations G(−133)A/C in CRE2 (46.7%), C(+160)T in DAS (30%) and A(310)del in BLV-mir-B4-5p, A(357)G in BLV-mir-B4-3p, A(462)G in BLV-mir-B5-5p, and GA(497–498)AG in BLV-mir-B5-3p (26.5%) were often seen in isolates with high PL and did not occur in the low PL group. In conclusion, we found several significant polymorphisms among BLV genomic sequences in Russia that would explain a progression towards higher or lower lymphoproliferation. The data presented in this article enabled the classification between two different genotypes; however, clear association between genotypes and the PL development was not found.
Full article
►▼
Show Figures
Open AccessReview
Current Developments in the Epidemiology and Control of Enzootic Bovine Leukosis as Caused by Bovine Leukemia Virus
by
Paul C. Bartlett, Vickie J. Ruggiero, Holden C. Hutchinson, Casey J. Droscha, Bo Norby, Kelly R. B. Sporer and Tasia M. Taxis
Cited by 30 | Viewed by 4713
Abstract
Enzootic Bovine Leukosis (EBL) caused by the bovine leukemia virus (BLV) has been eradicated in over 20 countries. In contrast, the U.S. and many other nations are experiencing increasing prevalence in the absence of efforts to control transmission. Recent studies have shown that
[...] Read more.
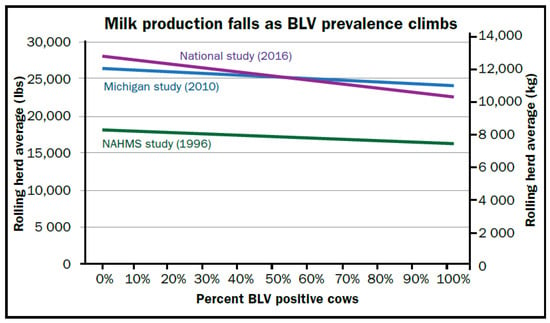
Enzootic Bovine Leukosis (EBL) caused by the bovine leukemia virus (BLV) has been eradicated in over 20 countries. In contrast, the U.S. and many other nations are experiencing increasing prevalence in the absence of efforts to control transmission. Recent studies have shown that BLV infection in dairy cattle has a greater impact beyond the long-recognized lymphoma development that occurs in <5% of infected cattle. Like other retroviruses, BLV appears to cause multiple immune system disruptions, affecting both cellular and humoral immunity, which are likely responsible for increasingly documented associations with decreased dairy production and decreased productive lifespan. Realization of these economic losses has increased interest in controlling BLV using technology that was unavailable decades ago, when many nations eradicated BLV via traditional antibody testing and slaughter methods. This traditional control is not economically feasible for many nations where the average herd antibody prevalence is rapidly approaching 50%. The ELISA screening of cattle with follow-up testing via qPCR for proviral load helps prioritize the most infectious cattle for segregation or culling. The efficacy of this approach has been demonstrated in at least four herds. Breeding cattle for resistance to BLV disease progression also appears to hold promise, and several laboratories are working on BLV vaccines. There are many research priorities for a wide variety of disciplines, especially including the need to investigate the reports linking BLV and human breast cancer.
Full article
►▼
Show Figures
Open AccessArticle
Bovine Leukemia Virus Infection Affects Host Gene Expression Associated with DNA Mismatch Repair
by
Lanlan Bai, Tomoya Hirose, Wlaa Assi, Satoshi Wada, Shin-nosuke Takeshima and Yoko Aida
Cited by 9 | Viewed by 2743
Abstract
Bovine leukemia virus (BLV) causes enzootic bovine leukosis, a malignant form of B-cell lymphoma, and is closely related to human T-cell leukemia viruses. We investigated whether BLV infection affects host genes associated with DNA mismatch repair (MMR). Next-generation sequencing of blood samples from
[...] Read more.
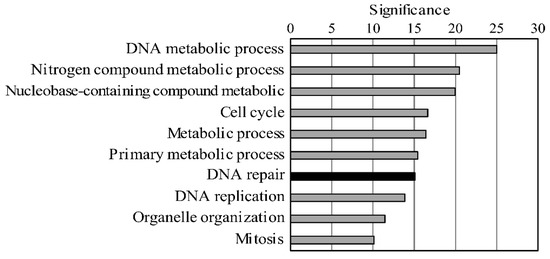
Bovine leukemia virus (BLV) causes enzootic bovine leukosis, a malignant form of B-cell lymphoma, and is closely related to human T-cell leukemia viruses. We investigated whether BLV infection affects host genes associated with DNA mismatch repair (MMR). Next-generation sequencing of blood samples from five calves experimentally infected with BLV revealed the highest expression levels of seven MMR genes (
EXO1,
UNG,
PCNA,
MSH2,
MSH3,
MSH6, and
PMS2) at the point of peak proviral loads (PVLs). Furthermore, MMR gene expression was only upregulated in cattle with higher PVLs. In particular, the expression levels of
MSH2,
MSH3, and
UNG positively correlated with PVL in vivo. The expression levels of all seven MMR genes in pig kidney-15 cells and the levels of
PMS2 and
EXO1 in HeLa cells also increased tendencies after transient transfection with a BLV infectious clone. Moreover, MMR gene expression levels were significantly higher in BLV-expressing cell lines compared with those in the respective parental cell lines. Expression levels of
MSH2 and
EXO1 in BLV-infected cattle with lymphoma were significantly lower and higher, respectively, compared with those in infected cattle in vivo. These results reveal that BLV infection affects MMR gene expression, offering new candidate markers for lymphoma diagnosis.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceArticle
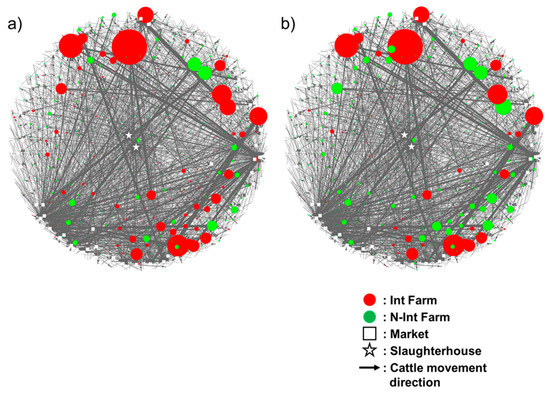
Quantitative Risk Assessment for the Introduction of Bovine Leukemia Virus-Infected Cattle Using a Cattle Movement Network Analysis
by
Kosuke Notsu, Anuwat Wiratsudakul, Shuya Mitoma, Hala El Daous, Chiho Kaneko, Heba M. El-Khaiat, Junzo Norimine and Satoshi Sekiguchi
Cited by 6 | Viewed by 2335
Abstract
The cattle industry is suffering economic losses caused by bovine leukemia virus (BLV) and enzootic bovine leukosis (EBL), the clinical condition associated with BLV infection. This pathogen spreads easily without detection by farmers and veterinarians due to the lack of obvious clinical signs.
[...] Read more.
The cattle industry is suffering economic losses caused by bovine leukemia virus (BLV) and enzootic bovine leukosis (EBL), the clinical condition associated with BLV infection. This pathogen spreads easily without detection by farmers and veterinarians due to the lack of obvious clinical signs. Cattle movement strongly contributes to the inter-farm transmission of BLV. This study quantified the farm-level risk of BLV introduction using a cattle movement analysis. A generalized linear mixed model predicting the proportion of BLV-infected cattle was constructed based on weighted in-degree centrality. Our results suggest a positive association between weighted in-degree centrality and the estimated number of introduced BLV-infected cattle. Remarkably, the introduction of approximately six cattle allowed at least one BLV-infected animal to be added to the farm in the worst-case scenario. These data suggest a high risk of BLV infection on farms with a high number of cattle being introduced. Our findings indicate the need to strengthen BLV control strategies, especially along the chain of cattle movement.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceArticle
Effects of Naturally Occurring Mutations in Bovine Leukemia Virus 5′-LTR and Tax Gene on Viral Transcriptional Activity
by
Aneta Pluta, Luc Willems, Renée N. Douville and Jacek Kuźmak
Cited by 12 | Viewed by 2675
Abstract
Bovine leukemia virus (BLV) is a deltaretrovirus infecting bovine B cells and causing enzootic bovine leucosis (EBL). The long terminal repeat (LTR) plays an indispensable role in viral gene expression. The BLV Tax protein acts as the main transactivator of LTR-driven transcription of
[...] Read more.
Bovine leukemia virus (BLV) is a deltaretrovirus infecting bovine B cells and causing enzootic bovine leucosis (EBL). The long terminal repeat (LTR) plays an indispensable role in viral gene expression. The BLV Tax protein acts as the main transactivator of LTR-driven transcription of BLV viral genes. The aim of this study was to analyze mutations in the BLV LTR region and
tax gene to determine their association with transcriptional activity. LTRs were obtained from one hundred and six BLV isolates and analyzed for their genetic variability. Fifteen variants were selected and characterized based on mutations in LTR regulatory elements, and further used for in vitro transcription assays. Reporter vectors containing the luciferase gene under the control of each variant BLV promoter sequence, in addition to variant Tax expression vectors, were constructed. Both types of plasmids were used for cotransfection of HeLa cells and the level of luciferase activity was measured as a proxy of transcriptional activity. Marked differences in LTR promoter activity and Tax transactivation activity were observed amongst BLV variants. These results demonstrate that mutations in both the BLV LTR and
tax gene can affect the promoter activity, which may have important consequences on proviral load, viral fitness, and transmissibility in BLV-infected cattle.
Full article
►▼
Show Figures