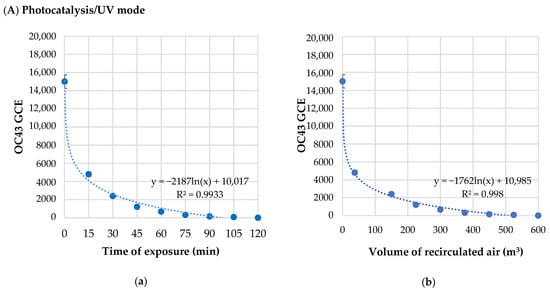
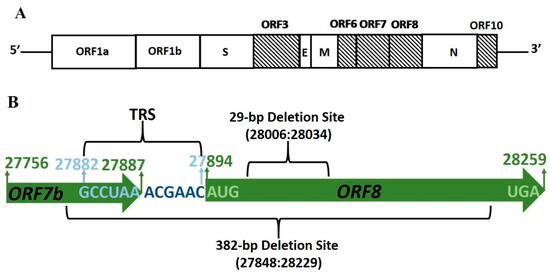
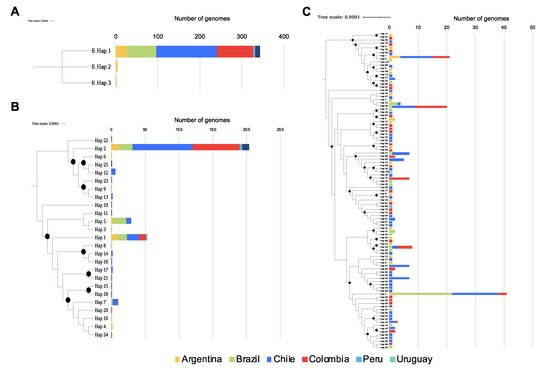
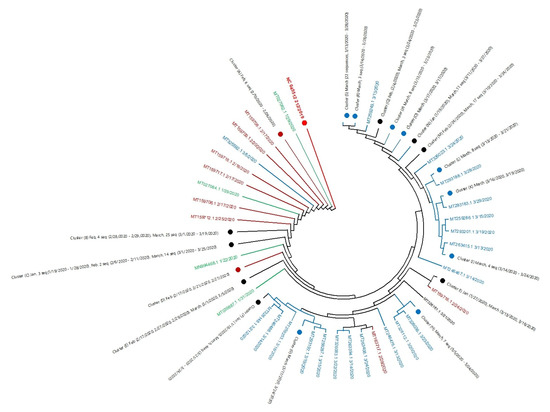
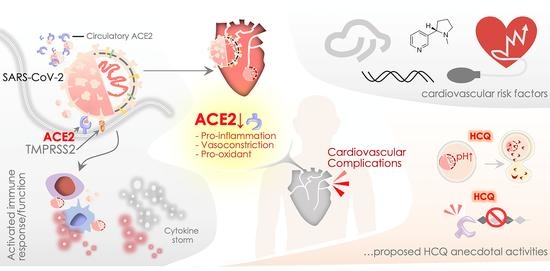
SARS-CoV Infections
A topical collection in Pathogens (ISSN 2076-0817). This collection belongs to the section "Viral Pathogens".
Viewed by 485982Editor
Topical Collection Information
Dear Colleagues,
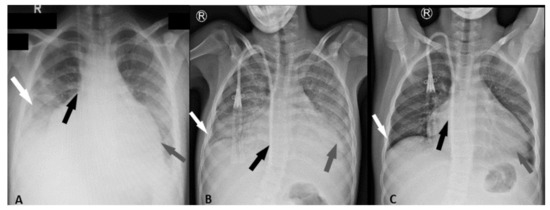
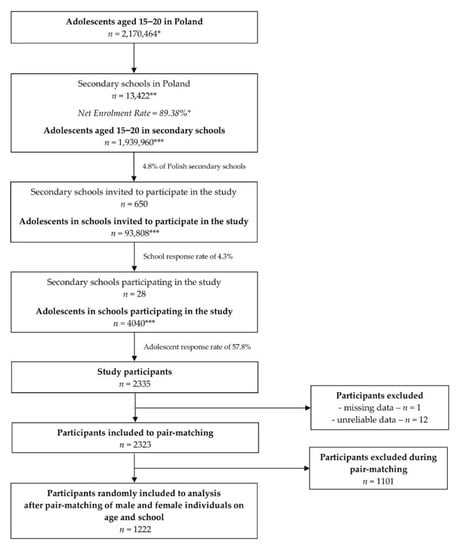
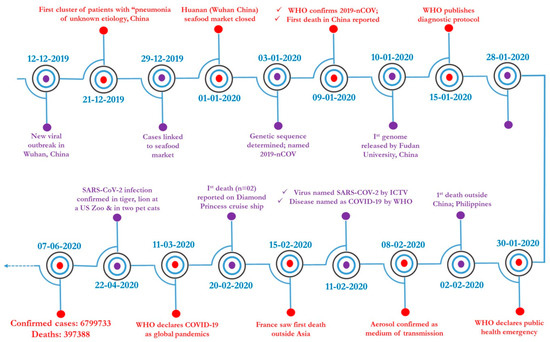
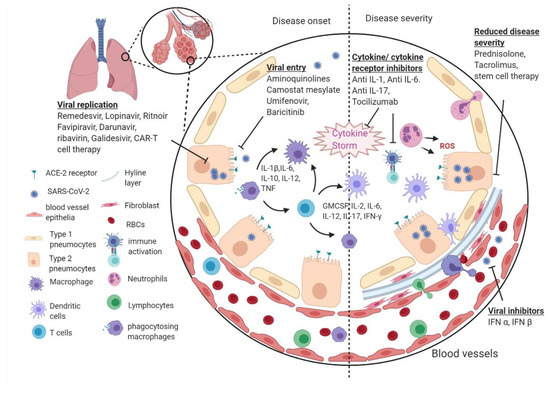
SARS Coronavirus (CoV) caused the first epidemic of the 21st century. The zoonotic emergence of SARS-CoV highlights the risk of rapid global spread of a highly infectious, novel pathogen. Disease ranged from mild respiratory symptoms to development of acute respiratory distress syndrome (ARDS) and death; ultimately the virus infected over 8,000 people with a 10% mortality rate. While SARS-CoV was contained by effective public health measures, a number of highly related zoonotic viruses have been found in bats that are fully capable of infecting human cells.
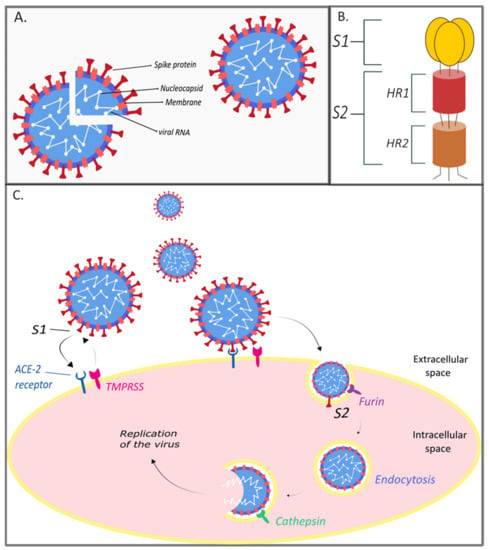
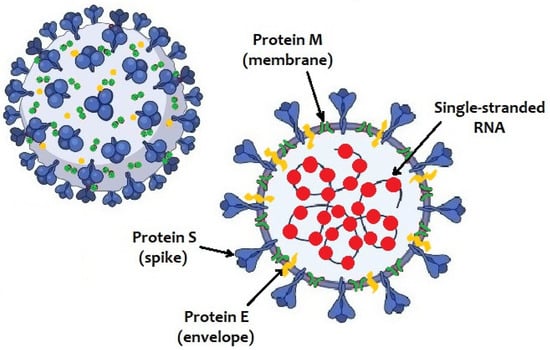
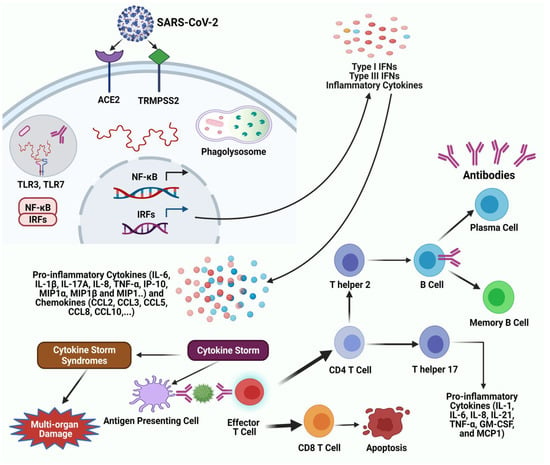
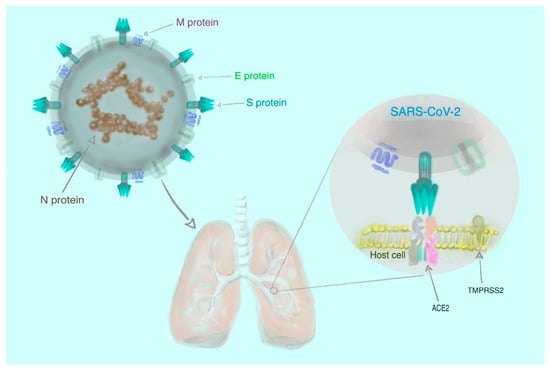
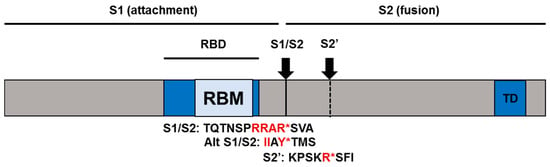
In the 16 years since SARS-CoV was identified, much has been learned about its emergence, replication and resulting disease. Several reverse genetics systems have allowed for investigation of the role of viral genes in both in vitro and in vivo infections while development of small animal models has allowed examination of the role of the host immune response following infection. Recent advances in structural biology have provided critical insights into how the viral spike protein binds to its cellular receptor, Ace2.
We would like to invite you to contribute to a Topical Collection on SARS-CoV Infections. In this Special Issue we welcome the coronavirus community to submit a research article, review or short communication focused on SARS-CoV emergence, replication, disease pathogenesis and protection, host immune response modulation, structural insights or anti-viral therapeutics.
We look forward to your contribution.
Dr. Lisa Gralinski
Collection Editor
Circulating bat viruses and emerging coronaviruses, recent structure advances, reverse genetics systems, immune mediated disease, 1st epidemic of 21st century
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Pathogens is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- coronavirus
- SARS-CoV
- respiratory virus
- viral pathogenesis
- immune response
- zoonosis