New Frontiers in Nucleic Acid Chemistry
A topical collection in Molecules (ISSN 1420-3049). This collection belongs to the section "Chemical Biology".
Viewed by 76731
Share This Topical Collection
Editors
 Prof. Dr. Ramon Eritja
Prof. Dr. Ramon Eritja
 Prof. Dr. Ramon Eritja
Prof. Dr. Ramon Eritja
E-Mail
Website
Collection Editor
1. Institute for Advanced Chemistry of Catalonia (IQAC-CSIC), Jordi Girona 18-26, E-08034 Barcelona, Spain
2. Networking Center on Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), Jordi Girona 18-26, E-08034 Barcelona, Spain
Interests: siRNA; antisense oligonucleotides; G-quadruplex; DNA triplex; i-motif; DNA nanotechnology; gene silencing; aptamers; drug delivery; DNA; RNA; oligonucleotide synthesis
Special Issues, Collections and Topics in MDPI journals
 Dr. Lajos Kovács
Dr. Lajos Kovács
 Dr. Lajos Kovács
Dr. Lajos Kovács
E-Mail
Website
Collection Editor
Nucleic Acids Laboratory, Department of Medicinal Chemistry, University of Szeged, Dóm tér 8, H-6720 Szeged, Hungary
Interests: G quadruplexes; supramolecular chemistry; synthetic organic chemistry of carbohydrates; nucleobases; nucleosides; C-nucleosides; peptide nucleic acids; heterocycles; protecting groups
Special Issues, Collections and Topics in MDPI journals
 Prof. Dr. Daniela Montesarchio
Prof. Dr. Daniela Montesarchio
 Prof. Dr. Daniela Montesarchio
Prof. Dr. Daniela Montesarchio
E-Mail
Website
Collection Editor
Department of Chemical Sciences, University of Naples Federico II, via Cintia 4, 80126 Naples, Italy
Interests: oligonucleotides; nucleoside analogues; unusual structures of DNA; G-quadruplex; Ru(III)-complexes; Pt(II)-complexes; metal–DNA interactions; small molecule–DNA interactions
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
After the success of the Special Issues “Nucleic Acid Analogs”, edited by Molecules in 2012, and “Frontiers in Nucleic Acid Chemistry”, edited in 2015, we are happy to announce the launch of a Topical Collection on these topics. Synthetic oligonucleotides have become essential tools for biological, biomedical, and nanotechnology researches and have also shown promising results in therapeutics and diagnostics. Novel oligonucleotide derivatives with tailored properties are being continuously developed since their specific recognition and unique folding characteristics allow to obtain potential drugs or drug delivery agents, biosensors, diagnostic agents, etc. Along with this development, nucleic acid research is taking new directions as well: synthesis and applications of even more complex molecules, like synthetic RNA analogues (peptide nucleic acids, locked nucleic acids, morpholino oligos, conjugates, etc.), highly informative analytical methods (e.g., those based on mass spectrometry), study of higher-order structures (triple and quadruple helices), investigation of molecular electronic devices based on smart, self-assembling oligonucleotide analogues, to name just a few. The study of the properties of modified nucleic acids brings together many different branches of science, such as molecular biology, chemistry, biochemistry, medicinal chemistry, medicine, materials science, and synthetic biology.
The Topical Collection of Molecules, “New Frontiers in Nucleic Acid Chemistry”, will concentrate on the latest developments in nucleic acids chemistry. We cordially invite all the researchers involved in this exciting field to contribute to the continuing success of the Topical Collection “New Frontiers in Nucleic Acid Chemistry”.
Prof. Dr. Ramon Eritja
Dr. Lajos Kovács
Prof. Dr. Daniela Montesarchio
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Molecules is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- modified oligonucleotides
- artificial nucleic acids
- xeno nucleic acids
- synthetic genes
- oligonucleotide conjugates
- nucleoside/nucleotide analogs
- G-quadruplex
- i-motif
- triplex
- antisense
- siRNA
- RNA interference
- antiviral activity
- anticancer activity
- DNA nanobiotechnology
- nanomaterials
- aptamers
- biosensors
- surface-plasmon resonance
- fluorescent nucleic acids
- locked nucleic acids
- unlocked nucleic acids
- peptide nucleic acids
- DNA repair
- base-modified DNA
- DNA polymerase
- circular nucleic acids
- NMR of nucleic acids
- mass spectrometry of nucleic acids
- biophysical studies
Related Special Issues
Published Papers (13 papers)
Open AccessReview
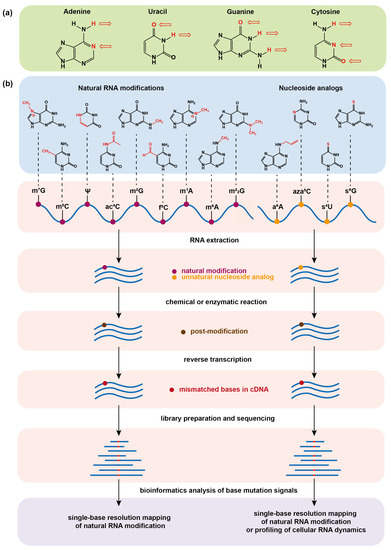
Determining RNA Natural Modifications and Nucleoside Analog-Labeled Sites by a Chemical/Enzyme-Induced Base Mutation Principle
by
Ziming Bao, Tengwei Li and Jianzhao Liu
Cited by 3 | Viewed by 1957
Abstract
The natural chemical modifications of messenger RNA (mRNA) in living organisms have shown essential roles in both physiology and pathology. The mapping of mRNA modifications is critical for interpreting their biological functions. In another dimension, the synthesized nucleoside analogs can enable chemical labeling
[...] Read more.
The natural chemical modifications of messenger RNA (mRNA) in living organisms have shown essential roles in both physiology and pathology. The mapping of mRNA modifications is critical for interpreting their biological functions. In another dimension, the synthesized nucleoside analogs can enable chemical labeling of cellular mRNA through a metabolic pathway, which facilitates the study of RNA dynamics in a pulse-chase manner. In this regard, the sequencing tools for mapping both natural modifications and nucleoside tags on mRNA at single base resolution are highly necessary. In this work, we review the progress of chemical sequencing technology for determining both a variety of naturally occurring base modifications mainly on mRNA and a few on transfer RNA and metabolically incorporated artificial base analogs on mRNA, and further discuss the problems and prospects in the field.
Full article
►▼
Show Figures
Open AccessArticle
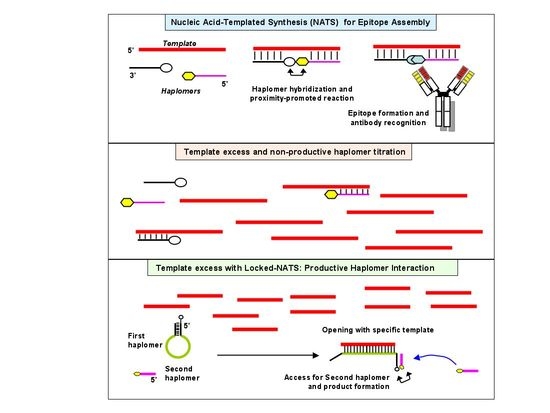
Assembly of Biologically Functional Structures by Nucleic Acid Templating: Implementation of a Strategy to Overcome Inhibition by Template Excess
by
Matthew M. Lawler, James T. Kurnick, Leah Fagundes St. Pierre, Estelle E. Newton, Lenora B. Rose and Ian S. Dunn
Viewed by 1286
Abstract
Delivery of therapeutic molecules to pathogenic cells is often hampered by unintended toxicity to normal cells. In principle, this problem can be circumvented if the therapeutic effector molecule is split into two inactive components, and only assembled on or within the target cell
[...] Read more.
Delivery of therapeutic molecules to pathogenic cells is often hampered by unintended toxicity to normal cells. In principle, this problem can be circumvented if the therapeutic effector molecule is split into two inactive components, and only assembled on or within the target cell itself. Such an in situ process can be realized by exploiting target-specific molecules as templates to direct proximity-enhanced assembly. Modified nucleic acids carrying inert precursor fragments can be designed to co-hybridize on a target-specific template nucleic acid, such that the enforced proximity accelerates assembly of a functional molecule for antibody recognition. We demonstrate the in vitro feasibility of this adaptation of nucleic acid-templated synthesis (NATS) using oligonucleotides bearing modified peptides (“haplomers”), for templated assembly of a mimotope recognized by the therapeutic antibody trastuzumab. Enforced proximity promotes mimotope assembly via traceless native chemical ligation. Nevertheless, titration of participating haplomers through template excess is a potential limitation of trimolecular NATS. In order to overcome this problem, we devised a strategy where haplomer hybridization can only occur in the presence of target, without being subject to titration effects. This generalizable NATS modification may find future applications in enabling directed targeting of pathological cells.
Full article
►▼
Show Figures
Open AccessArticle
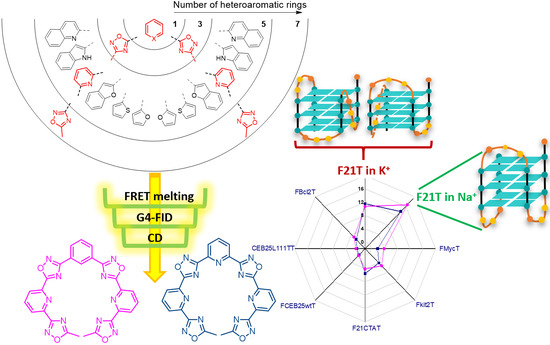
Oxadiazole/Pyridine-Based Ligands: A Structural Tuning for Enhancing G-Quadruplex Binding
by
Filippo Doria, Valentina Pirota, Michele Petenzi, Marie-Paule Teulade-Fichou, Daniela Verga and Mauro Freccero
Cited by 16 | Viewed by 4237
Abstract
Non-macrocyclic heteroaryls represent a valuable class of ligands for nucleic acid recognition. In this regard, non-macrocyclic pyridyl polyoxazoles and polyoxadiazoles were recently identified as selective G-quadruplex stabilizing compounds with high cytotoxicity and promising anticancer activity. Herein, we describe the synthesis of a new
[...] Read more.
Non-macrocyclic heteroaryls represent a valuable class of ligands for nucleic acid recognition. In this regard, non-macrocyclic pyridyl polyoxazoles and polyoxadiazoles were recently identified as selective G-quadruplex stabilizing compounds with high cytotoxicity and promising anticancer activity. Herein, we describe the synthesis of a new family of heteroaryls containing oxadiazole and pyridine moieties targeting DNA G-quadruplexes. To perform a structure–activity analysis identifying determinants of activity and selectivity, we followed a convergent synthetic pathway to modulate the nature and number of the heterocycles (1,3-oxazole vs. 1,2,4-oxadiazole and pyridine vs. benzene). Each ligand was evaluated towards secondary nucleic acid structures, which have been chosen as a prototype to mimic cancer-associated G-quadruplex structures (e.g., the human telomeric sequence, c-myc and c-kit promoters). Interestingly, heptapyridyl-oxadiazole compounds showed preferential binding towards the telomeric sequence (22AG) in competitive conditions vs. duplex DNA. In addition, G4-FID assays suggest a different binding mode from the classical stacking on the external G-quartet. Additionally, CD titrations in the presence of the two most promising compounds for affinity, TOxAzaPy and TOxAzaPhen, display a structural transition of 22AG in K-rich buffer. This investigation suggests that the pyridyl-oxadiazole motif is a promising recognition element for G-quadruplexes, combining seven heteroaryls in a single binding unit.
Full article
►▼
Show Figures
Open AccessArticle
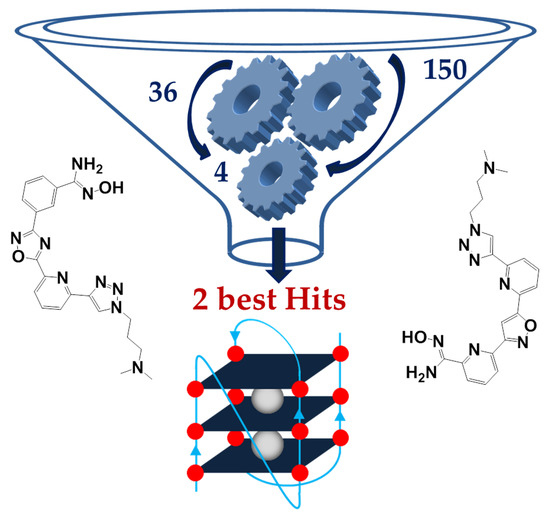
A Fragment-Based Approach for the Development of G-Quadruplex Ligands: Role of the Amidoxime Moiety
by
Martina Tassinari, Alberto Lena, Elena Butovskaya, Valentina Pirota, Matteo Nadai, Mauro Freccero, Filippo Doria and Sara N. Richter
Cited by 11 | Viewed by 5010
Abstract
G-quadruplex (G4) nucleic acid structures have been reported to be involved in several human pathologies, including cancer, neurodegenerative disorders and infectious diseases; however, G4 targeting compounds still need implementation in terms of drug-like properties and selectivity in order to reach the clinical use.
[...] Read more.
G-quadruplex (G4) nucleic acid structures have been reported to be involved in several human pathologies, including cancer, neurodegenerative disorders and infectious diseases; however, G4 targeting compounds still need implementation in terms of drug-like properties and selectivity in order to reach the clinical use. So far, G4 ligands have been mainly identified through high-throughput screening methods or design of molecules with pre-set features. Here, we describe the development of new heterocyclic ligands through a fragment-based drug discovery (FBDD) approach. The ligands were designed against the major G4 present in the long terminal repeat (LTR) promoter region of the human immunodeficiency virus-1 (HIV-1), the stabilization of which has been shown to suppress viral gene expression and replication. Our method is based on the generation of molecular fragment small libraries, screened against the target to further elaborate them into lead compounds. We screened 150 small molecules, composed by structurally and chemically different fragments, selected from commercially available and in-house compounds; synthetic elaboration yielded several G4 ligands and two final G4 binders, both embedding an amidoxime moiety; one of these two compounds showed preferential binding for the HIV-1 LTR G4. This work presents the discovery of a novel potential pharmacophore and highlights the possibility to apply a fragment-based approach to develop G4 ligands with unexpected chemical features.
Full article
►▼
Show Figures
Open AccessArticle
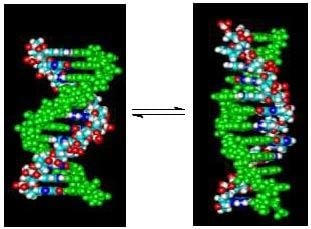
Linking Temperature, Cation Concentration and Water Activity for the B to Z Conformational Transition in DNA
by
Jaime M. Ferreira and Richard D. Sheardy
Cited by 7 | Viewed by 4285
Abstract
High concentrations of Na
+ or [Co(NH
3)
6]
3+ can induce the B to Z conformational transition in alternating (dC-dG) oligo and polynucleotides. The use of short DNA oligomers (dC-dG)
4 and (dm
5C-dG)
4 as models can allow
[...] Read more.
High concentrations of Na
+ or [Co(NH
3)
6]
3+ can induce the B to Z conformational transition in alternating (dC-dG) oligo and polynucleotides. The use of short DNA oligomers (dC-dG)
4 and (dm
5C-dG)
4 as models can allow a thermodynamic characterization of the transition. Both form right handed double helical structures (B-DNA) in standard phosphate buffer with 115 mM Na
+ at 25 °C. However, at 2.0 M Na
+ or 200 μM [Co(NH
3)
6]
3+, (dm
5C-dG)
4 assumes a left handed double helical structure (Z-DNA) while the unmethylated (dC-dG)
4 analogue remains right handed under those conditions. We have previously demonstrated that the enthalpy of the transition at 25 °C for either inducer can be determined using isothermal titration calorimetry (ITC). Here, ITC is used to investigate the linkages between temperature, water activity and DNA conformation. We found that the determined enthalpy for each titration varied linearly with temperature allowing determination of the heat capacity change (ΔC
p) between the initial and final states. As expected, the ΔC
p values were dependent upon the cation (i.e., Na
+ vs. [Co(NH
3)
6]
3+) as well as the sequence of the DNA oligomer (i.e., methylated vs. unmethylated). Osmotic stress experiments were carried out to determine the gain or loss of water by the oligomer induced by the titration. The results are discussed in terms of solvent accessible surface areas, electrostatic interactions and the role of water.
Full article
►▼
Show Figures
Open AccessReview
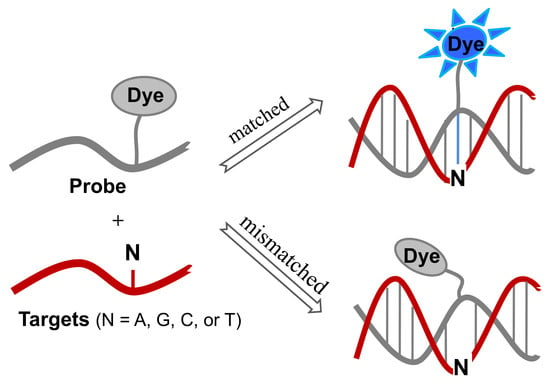
Single-Labeled Oligonucleotides Showing Fluorescence Changes upon Hybridization with Target Nucleic Acids
by
Gil Tae Hwang
Cited by 23 | Viewed by 7967
Abstract
Sequence-specific detection of nucleic acids has been intensively studied in the field of molecular diagnostics. In particular, the detection and analysis of single-nucleotide polymorphisms (SNPs) is crucial for the identification of disease-causing genes and diagnosis of diseases. Sequence-specific hybridization probes, such as molecular
[...] Read more.
Sequence-specific detection of nucleic acids has been intensively studied in the field of molecular diagnostics. In particular, the detection and analysis of single-nucleotide polymorphisms (SNPs) is crucial for the identification of disease-causing genes and diagnosis of diseases. Sequence-specific hybridization probes, such as molecular beacons bearing the fluorophore and quencher at both ends of the stem, have been developed to enable DNA mutation detection. Interestingly, DNA mutations can be detected using fluorescently labeled oligonucleotide probes with only one fluorophore. This review summarizes recent research on single-labeled oligonucleotide probes that exhibit fluorescence changes after encountering target nucleic acids, such as guanine-quenching probes, cyanine-containing probes, probes containing a fluorophore-labeled base, and microenvironment-sensitive probes.
Full article
►▼
Show Figures
Open AccessFeature PaperReview
Recent Advances in Nucleic Acid Targeting Probes and Supramolecular Constructs Based on Pyrene-Modified Oligonucleotides
by
Olga A. Krasheninina, Darya S. Novopashina, Evgeny K. Apartsin and Alya G. Venyaminova
Cited by 36 | Viewed by 9790
Abstract
In this review, we summarize the recent advances in the use of pyrene-modified oligonucleotides as a platform for functional nucleic acid-based constructs. Pyrene is of special interest for the development of nucleic acid-based tools due to its unique fluorescent properties (sensitivity of fluorescence
[...] Read more.
In this review, we summarize the recent advances in the use of pyrene-modified oligonucleotides as a platform for functional nucleic acid-based constructs. Pyrene is of special interest for the development of nucleic acid-based tools due to its unique fluorescent properties (sensitivity of fluorescence to the microenvironment, ability to form excimers and exciplexes, long fluorescence lifetime, high quantum yield), ability to intercalate into the nucleic acid duplex, to act as a π-π-stacking (including anchoring) moiety, and others. These properties of pyrene have been used to construct novel sensitive fluorescent probes for the sequence-specific detection of nucleic acids and the discrimination of single nucleotide polymorphisms (SNPs), aptamer-based biosensors, agents for binding of double-stranded DNAs, and building blocks for supramolecular complexes. Special attention is paid to the influence of the design of pyrene-modified oligonucleotides on their properties, i.e., the structure-function relationships. The perspectives for the applications of pyrene-modified oligonucleotides in biomolecular studies, diagnostics, and nanotechnology are discussed.
Full article
►▼
Show Figures
Open AccessArticle
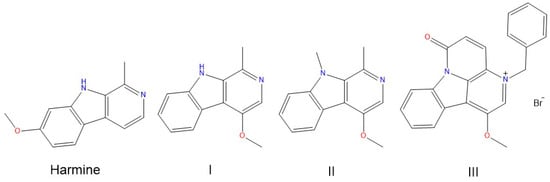
Binding of Harmine Derivatives to DNA: A Spectroscopic Investigation
by
Bruno Pagano, Marco Caterino, Rosanna Filosa and Concetta Giancola
Cited by 12 | Viewed by 5140
Abstract
Harmine belongs to a group of β-carboline alkaloids endowed with antitumor properties. Harmine and its derivatives are thought to bind to DNA and interfere with topoisomerase activities. We investigated the base-dependent binding of harmine, and three of its synthetic anticancer-active derivatives to the
[...] Read more.
Harmine belongs to a group of β-carboline alkaloids endowed with antitumor properties. Harmine and its derivatives are thought to bind to DNA and interfere with topoisomerase activities. We investigated the base-dependent binding of harmine, and three of its synthetic anticancer-active derivatives to the genomic DNA from calf thymus and two synthetic 20-mer double helices, the poly(dG-dC)·poly(dG-dC) and the poly(dA-dT)·poly(dA-dT), by means of UV-Vis and circular dichroism (CD) spectroscopies. The data show that the DNA binding and stabilising properties of the investigated derivatives are base pair-dependent. These results could be used as a guide to design and develop further bioactive analogues.
Full article
►▼
Show Figures
Open AccessArticle
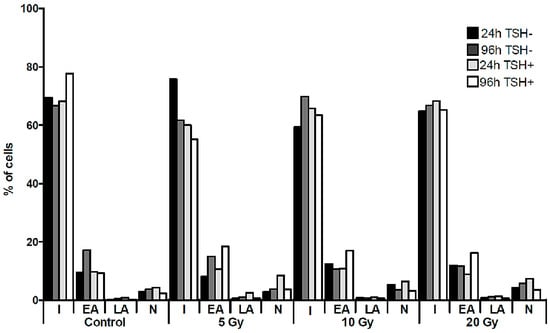
The Molecular Effect of Diagnostic Absorbed Doses from 131I on Papillary Thyroid Cancer Cells In Vitro
by
Mariusz Stasiołek, Zbigniew Adamczewski, Przemysław W. Śliwka, Bartosz Puła, Bolesław Karwowski, Anna Merecz-Sadowska, Marek Dedecjus and Andrzej Lewiński
Cited by 5 | Viewed by 4539
Abstract
Diagnostic whole-body scan is a standard procedure in patients with thyroid cancer prior to the application of a therapeutic dose of
131I. Unfortunately, administration of the radioisotope in a diagnostic dose may decrease further radioiodine uptake—the phenomenon called “thyroid stunning”. We estimated
[...] Read more.
Diagnostic whole-body scan is a standard procedure in patients with thyroid cancer prior to the application of a therapeutic dose of
131I. Unfortunately, administration of the radioisotope in a diagnostic dose may decrease further radioiodine uptake—the phenomenon called “thyroid stunning”. We estimated radiation absorbed dose-dependent changes in genetic material, in particular in the sodium iodide symporter (NIS) gene promoter, and the NIS protein level in a K1 cell line derived from the metastasis of a human papillary thyroid carcinoma exposed to
131I in culture. The different activities applied were calculated to result in absorbed doses of 5, 10 and 20 Gy. Radioiodine did not affect the expression of the
NIS gene at the mRNA level, however, we observed significant changes in the NIS protein level in K1 cells. The decrease of the NIS protein level observed in the cells subjected to the lowest absorbed dose was paralleled by a significant increase in 8-oxo-dG concentrations (
p < 0.01) and followed by late activation of the DNA repair pathways. Our findings suggest that the impact of
131I radiation on thyroid cells, in the range compared to doses absorbed during diagnostic procedures, is not linear and depends on various factors including the cellular components of thyroid pathology.
Full article
►▼
Show Figures
Open AccessReview
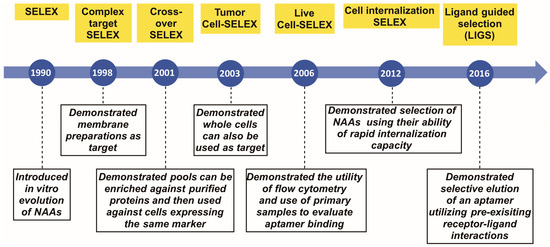
Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens
by
Prabodhika Mallikaratchy
Cited by 74 | Viewed by 9459
Abstract
The demand has increased for sophisticated molecular tools with improved detection limits. Such molecules should be simple in structure, yet stable enough for clinical applications. Nucleic acid aptamers (NAAs) represent a class of molecules able to meet this demand. In particular, aptamers, a
[...] Read more.
The demand has increased for sophisticated molecular tools with improved detection limits. Such molecules should be simple in structure, yet stable enough for clinical applications. Nucleic acid aptamers (NAAs) represent a class of molecules able to meet this demand. In particular, aptamers, a class of small nucleic acid ligands that are composed of single-stranded modified/unmodified RNA/DNA molecules, can be evolved from a complex library using Systematic Evolution of Ligands by EXponential enrichment (SELEX) against almost any molecule. Since its introduction in 1990, in stages, SELEX technology has itself undergone several modifications, improving selection and broadening the repertoire of targets. This review summarizes these milestones that have pushed the field forward, allowing researchers to generate aptamers that can potentially be applied as therapeutic and diagnostic agents.
Full article
►▼
Show Figures
Open AccessArticle
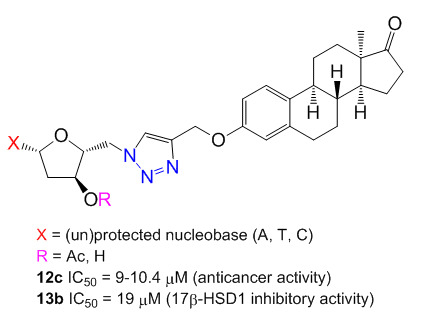
Synthesis and Biological Evaluation of Triazolyl 13α-Estrone–Nucleoside Bioconjugates
by
Brigitta Bodnár, Erzsébet Mernyák, János Wölfling, Gyula Schneider, Bianka Edina Herman, Mihály Szécsi, Izabella Sinka, István Zupkó, Zoltán Kupihár and Lajos Kovács
Cited by 16 | Viewed by 7091
Abstract
2′-Deoxynucleoside conjugates of 13α-estrone were synthesized by applying the copper-catalyzed alkyne–azide click reaction (CuAAC). For the introduction of the azido group the 5′-position of the nucleosides and a propargyl ether functional group on the 3-hydroxy group of 13α-estrone were chosen. The best yields
[...] Read more.
2′-Deoxynucleoside conjugates of 13α-estrone were synthesized by applying the copper-catalyzed alkyne–azide click reaction (CuAAC). For the introduction of the azido group the 5′-position of the nucleosides and a propargyl ether functional group on the 3-hydroxy group of 13α-estrone were chosen. The best yields were realized in our hands when the 3′-hydroxy groups of the nucleosides were protected by acetyl groups and the 5′-hydroxy groups were modified by the tosyl–azide exchange method. The commonly used conditions for click reaction between the protected-5′-azidonucleosides and the steroid alkyne was slightly modified by using 1.5 equivalent of Cu(I) catalyst. All the prepared conjugates were evaluated in vitro by means of MTT assays for antiproliferative activity against a panel of human adherent cell lines (HeLa, MCF-7 and A2780) and the potential inhibitory activity of the new conjugates on human 17β-hydroxysteroid dehydrogenase 1 (17β-HSD1) was investigated via in vitro radiosubstrate incubation. Some protected conjugates displayed moderate antiproliferative properties against a panel of human adherent cancer cell lines (the protected cytidine conjugate proved to be the most potent with IC
50 value of 9 μM). The thymidine conjugate displayed considerable 17β-HSD1 inhibitory activity (IC
50 = 19 μM).
Full article
►▼
Show Figures
Open AccessArticle
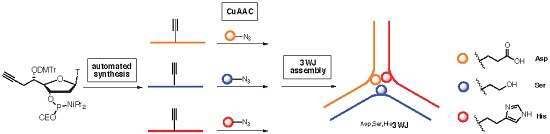
DNA Three Way Junction Core Decorated with Amino Acids-Like Residues-Synthesis and Characterization
by
Claudia Addamiano, Béatrice Gerland, Corinne Payrastre and Jean-Marc Escudier
Cited by 4 | Viewed by 7465
Abstract
Construction and physico-chemical behavior of DNA three way junction (3WJ) functionalized by protein-like residues (imidazole, alcohol and carboxylic acid) at unpaired positions at the core is described. One 5′-C(
S)-propargyl-thymidine nucleotide was specifically incorporated on each strand to react through a post
[...] Read more.
Construction and physico-chemical behavior of DNA three way junction (3WJ) functionalized by protein-like residues (imidazole, alcohol and carboxylic acid) at unpaired positions at the core is described. One 5′-C(
S)-propargyl-thymidine nucleotide was specifically incorporated on each strand to react through a post synthetic CuACC reaction with either protected imidazolyl-, hydroxyl- or carboxyl-azide. Structural impacts of 5′-C(
S)-functionalization were investigated to evaluate how 3WJ flexibility/stability is affected.
Full article
►▼
Show Figures
Open AccessArticle
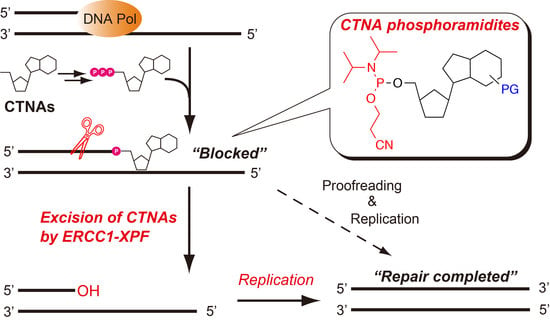
Chemical Incorporation of Chain-Terminating Nucleoside Analogs as 3′-Blocking DNA Damage and Their Removal by Human ERCC1-XPF Endonuclease
by
Junpei Yamamoto, Chiaki Takahata, Isao Kuraoka, Kouji Hirota and Shigenori Iwai
Cited by 3 | Viewed by 7092
Abstract
Nucleoside/nucleotide analogs that lack the 3′-hydroxy group are widely utilized for HIV therapy. These chain-terminating nucleoside analogs (CTNAs) block DNA synthesis after their incorporation into growing DNA, leading to the antiviral effects. However, they are also considered to be DNA damaging agents, and
[...] Read more.
Nucleoside/nucleotide analogs that lack the 3′-hydroxy group are widely utilized for HIV therapy. These chain-terminating nucleoside analogs (CTNAs) block DNA synthesis after their incorporation into growing DNA, leading to the antiviral effects. However, they are also considered to be DNA damaging agents, and tyrosyl-DNA phosphodiesterase 1, a DNA repair enzyme, is reportedly able to remove such CTNA-modifications of DNA. Here, we have synthesized phosphoramidite building blocks of representative CTNAs, such as acyclovir, abacavir, carbovir, and lamivudine, and oligonucleotides with the 3′-CTNAs were successfully synthesized on solid supports. Using the chemically synthesized oligonucleotides, we investigated the excision of the 3′-CTNAs in DNA by the human excision repair cross complementing protein 1-xeroderma pigmentosum group F (ERCC1-XPF) endonuclease, which is one of the main components of the nucleotide excision repair pathway. A biochemical analysis demonstrated that the ERCC1-XPF endonuclease cleaved 2–7 nt upstream from the 3′-blocking CTNAs, and that DNA synthesis by the Klenow fragment was resumed after the removal of the CTNAs, suggesting that ERCC1-XPF participates in the repair of the CTNA-induced DNA damage.
Full article
►▼
Show Figures
Planned Papers
The below list represents only planned manuscripts. Some of these
manuscripts have not been received by the Editorial Office yet. Papers
submitted to MDPI journals are subject to peer-review.
Title: Fragment-based expansion for the development of highly stabilizing HIV LTR G-quadruplex ligands
Author: Sara N. Richter
Affiliation: Department of Molecular Medicine, University of Padua, Italy
Email: sara.richter@unipd.it
Abstract: Stabilization of G-quadruplex (G4) structures in the HIV-1 LTR promoter region suppresses viral transcription, which suggests the use of selective G4 ligands as a novel anti-HIV therapeutic approach. However, sufficient selectivity for the viral versus the cellular G4s needs to be reached to adopt this strategy. Here, we describe the design of new heterocyclic ligands identified by a fragment-based drug discovery (FBDD) approach. Our method is based on the generation of molecular fragment small libraries, screened against the target to further elaborate them into lead compounds. We started with the screening of around 150 molecules, composed of structurally and chemically different fragments, selected from in-house libraries; further synthetic elaboration yielded several G4 ligands and two final potent and selective LTR G4 binders. This work highlights the possibility to apply a fragment-based approach to target G4s.
Title: Polyetheroaryl oxadiazole/pyridine-based ligands: a structural tuning for enhancing G-quadruplex binding
Author: Filippo Doria, Daniela Verga, Valentina Pirota, Marie-Paule Teulade-Fichou and Mauro Freccero
Affiliation: Università di Pavia, Dipartimento di Chimica , Pavia, Italy
Email: mauro.freccero@unipv.it
Abstract: Acyclic olygoheteroaryl-based compounds represent a valuable class of ligands for nucleic acid recognition. In this regard, acyclic pyridyl polyoxazoles and polyoxadiazoles were recently identified as selective G-quadruplex stabilizing compounds with high cytotoxicity and promising anticancer activity. Herein, we describe the synthesis of a new family of polyheteroaryl oxadiazole/pyridine-ligands targeting DNA G-quadruplexes. In order to perform a structure-activity analysis to identify determinants of activity and selectivity, we followed a convergent synthetic pathway to modulate the nature of the heterocycles, their number, and the effect of protonable lateral chains. Each ligand was evaluated towards secondary nucleic acid structures chosen as a prototype to mimic cancer-associated G-quadruplex structures (e.g., the human telomeric sequence, C-myc and C-kit promoters). Interesting, heptapyridyl-oxadiazole compounds showed preferential binding towards the telomeric sequence (22AG) and the C-myc oncogene promoter in competition conditions versus duplex DNA. In addition, G4-FID assays suggest a different binding mode from the classical stacking on the external G-quartet. Additionally, CD titrations in the presence of the two most promising compounds, TOxAzaPy and TOxAzaPhen, display a structural transition of 22AG in K-rich buffer. This investigation suggests that the pyridyl-oxadiazole motif is a promising recognition element for G-quadruplexes, combining seven heteroaryls in a single binding unit.
Title: Linking Temperature, Cation Concentration and Water Activity for the B to Z Conformational Transition in DNA
Author: Jaime M. Ferreira1# and Richard D. Sheardy2*
Affiliation: 1 Department of Chemistry and Biochemistry, Seton Hall University, South Orange, NJ 07079
2 Department of Chemistry and Biochemistry, Texas Woman’s University, Denton, TX 76204
Email: RSheardy@twu.edu
Abstract: High concentrations of Na+ or [Co(NH3)6]3+ can induce the B to Z conformational transition in alternating (dC-dG) oligo and polynucleotides. The use of short DNA oligomers (dC-dG)4 and (dm5C-dG)4 as models allows thermodynamic characterization of the transition. Both form right handed double helical structures in standard phosphate buffer with 115 mM Na+ at 25 oC. However, at 2.0 M Na+ or 200 μM [Co(NH3)6]3+, (dm5C-dG)4 assumes a left handed double helical structure while the unmethylated (dC-dG)4 analogue remains right handed. Here, isothermal titration calorimetry (ITC) is used to determine the linkages between temperature, cation concentration, water activity and the enthalpy of the conformational change. The results are discussed in terms of solvent accessible surface areas, electrostatic interactions and the role of water.