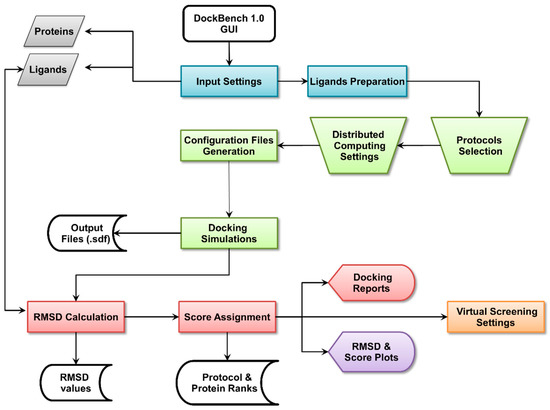
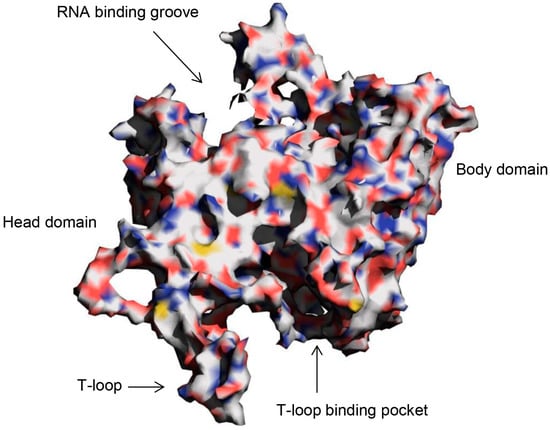
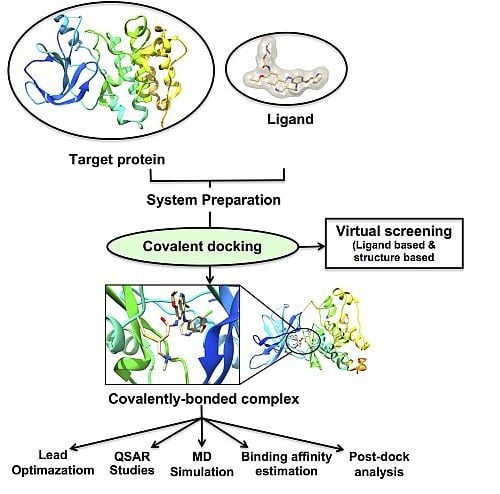
Molecular Docking (Closed)
A topical collection in Molecules (ISSN 1420-3049). This collection belongs to the section "Medicinal Chemistry".
Viewed by 82571Editors
Interests: medicinal chemistry; computational chemistry; 3D-QSAR; machine learning; drug design
Special Issues, Collections and Topics in MDPI journals
Interests: biochemistry; computer aided drug design; cheminformatics; bioinformatics
Special Issues, Collections and Topics in MDPI journals
Interests: Molecular Modeling of small molecules in transthyretin; G-Protein coupled receptors; metalloenzymes; Drug Design and ADMET in silico
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Following the success of the Special Issue entitled “Molecular Docking in Drug Design” ( https://www.mdpi.com/journal/molecules/special_issues/molecular_docking_drug ), where a number of papers have been published, it was decided to widen the issue and convert it to a Topical Collection under the name of “Molecular Docking”.
Molecular docking is an invaluable tool to study how two different molecular entities can recognize each to other. This tool is, not only used in modern drug discovery and design, but also in other fields, such as protein–protein interaction characterization to drive biological investigations.
Although many algorithms have been disclosed, and new docking programs are constantly released, there is still a lot to do. Many authors have proposed different scoring function approaches, such as consensus scoring, consensus docking and external independent scoring, others have proposed different poses generation methods or different conformation generators. Many methods have been reported to work better than others; nevertheless, we are still very far away from the ideal molecular docking software.
Therefore, there is still the need to continue the improvement of actual molecular docking procedures and this represent the main goal of this collection where manuscript will be accepted for publication if reporting newest approaches in molecular docking application and development.
The “Molecular Docking Topical Collection” will embrace several molecular docking related topics, such as:
- Small Molecule Reversible Docking Application
- Small Molecule Covalent Docking Application
- Virtual Screening
- De Novo Design
- Molecular Docking Software Development
- Molecular Docking Software Comparison
- Protein-Protein docking
- Scoring Function Development
- Scoring Function Comparison
- Drug Design
- Host-Guest Studies
- Molecular Dynamics as molecular Docking Tools
- Reviews
Dr. Rino Ragno
Dr. Milan Mladenovic
Dr. Gabriella Ortore
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Molecules is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Related Special Issue
- Molecular Docking in Drug Design in Molecules (15 articles - displayed below)