Antimicrobial Resistance
A topical collection in Life (ISSN 2075-1729). This collection belongs to the section "Microbiology".
Viewed by 58771
Share This Topical Collection
Editors
 Prof. Dr. Caterina Aurilio
Prof. Dr. Caterina Aurilio
 Prof. Dr. Caterina Aurilio
Prof. Dr. Caterina Aurilio
E-Mail
Website
Guest Editor
Anesthesia and Intensive Care Unit, University of Campania Luigi Vanvitelli, 81100 Naples, Italy
Interests: anaesthetics; pain medicine; anesthesiology; anesthesia; pain management and critical illness
 Dr. Antonella Paladini
Dr. Antonella Paladini
 Dr. Antonella Paladini
Dr. Antonella Paladini
E-Mail
Guest Editor
Anesthesiology and Pain Medicine, L'Aquila University, 67100 L'Aquila AQ, Italy
Interests: chronic pain; complementary medicine; low back pain; meditation; osteoarthritis; Pain
 Dr. Pasquale Sansone
Dr. Pasquale Sansone
 Dr. Pasquale Sansone
Dr. Pasquale Sansone
E-Mail
Website
Guest Editor
Department of Woman, Child and General and Specialized Surgery, University of Campania “Luigi Vanvitelli”, 81100 Naples, Italy
Interests: anesthesia, pain management, intensive care, pain treatment
 Dr. Vincenzo Pota
Dr. Vincenzo Pota
 Dr. Vincenzo Pota
Dr. Vincenzo Pota
E-Mail
Website
Guest Editor
Department of Woman, Child and General and Specialized Surgery, University of Campania “Luigi Vanvitelli”, 81100 Naples, Italy
Interests: intensive care unit; peri-operative medicine; postoperative pain especially opioid free management
Topical Collection Information
Dear Colleagues,
Antimicrobial resistance is a neverending problem, as many intensivists and physicians know. We are going to describe a wide range of conditions in which Antimicrobial Resistance represents a challenge and sometimes a pathological event difficult to resolve. This collection will comprise the topics below:
Antimicrobial resistance in intensive care
Blood stream infections from MDR bacteria
MDR pneumonia in intensive care
Multidrug resistance mycosis
Adiuvant therapy for MDR bacteria sepsis
MDR prevalence in COVID area
Emergent antibiotics for MDR
The antimicrobial stewardship in management of MDR infections
Dr. Caterina Aurilio
Dr. Antonella Paladini
Dr. Pasquale Sansone
Dr. Vincenzo Pota
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Life is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- Antimicrobial resistance
- stewardship
- Pneumonia
- Mycosis
- Bacteremia
- Sepsis
- Infections
- Antibiotics
Published Papers (16 papers)
Open AccessReview
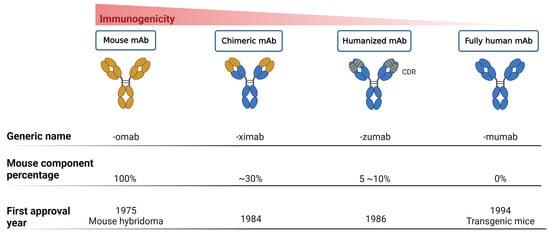
Monoclonal Antibodies as a Therapeutic Strategy against Multidrug-Resistant Bacterial Infections in a Post-COVID-19 Era
by
Hsiao-Chun Chen, Yu-Ling Pan, Ying Chen, Tsung-Hsuan Yang, Erh-Tung Hsu, Yu-Ting Huang and Ming-Hsien Chiang
Viewed by 1511
Abstract
The development of severe multidrug-resistant bacterial infections has recently intensified because of the COVID-19 pandemic. According to the guidelines issued by the World Health Organization (WHO), routine antibiotic administration is not recommended for patients with supposed or confirmed mild SARS-CoV-2 infection or pneumonia,
[...] Read more.
The development of severe multidrug-resistant bacterial infections has recently intensified because of the COVID-19 pandemic. According to the guidelines issued by the World Health Organization (WHO), routine antibiotic administration is not recommended for patients with supposed or confirmed mild SARS-CoV-2 infection or pneumonia, unless bacterial infection is clinically suspected. However, recent studies have pointed out that the proportion of non-essential antibiotic use in patients infected with SARS-CoV-2 remains high. Therefore, the silent pandemic of antibiotic resistance remains a pressing issue regardless of the present threats presented by the COVID-19 pandemic. To prevent or delay entry into the postulated post-antibiotic era, the long-term advocacy for the rational use of antibiotics, the optimization of infection control procedures, and the development of new antibacterial agents and vaccines should be underscored as vital practices of the antibacterial toolbox. Recently, the development of vaccines and monoclonal antibodies has gradually received attention following the advancement of biotechnology as well as enhanced drug discovery and development in cancer research. Although decent progress has been made in laboratory-based research and promising results have been obtained following clinical trials of some of these products, challenges still exist in their widespread clinical applications. This article describes the current advantages of antibacterial monoclonal antibodies, the development of associated clinical trials, and some perceived future perspectives and challenges. Further, we anticipate the development of more therapeutic agents to combat drug-resistant bacterial infections as well as to increase the resilience of current or novel agents/strategies.
Full article
►▼
Show Figures
Open AccessReview
Difficult-to-Treat Pathogens: A Review on the Management of Multidrug-Resistant Staphylococcus epidermidis
by
Valentina Siciliano, Rosa Anna Passerotto, Marta Chiuchiarelli, Gabriele Maria Leanza and Veronica Ojetti
Cited by 9 | Viewed by 4502
Abstract
Multidrug-resistant
Staphylococcus epidermidis (MDRSE) is responsible for difficult-to-treat infections in humans and hospital-acquired-infections. This review discusses the epidemiology, microbiology, diagnosis, and treatment of MDRSE infection and identifies knowledge gaps. By using the search term “pan resistant
Staphylococcus epidermidis” OR “multi-drug resistant
Staphylococcus
[...] Read more.
Multidrug-resistant
Staphylococcus epidermidis (MDRSE) is responsible for difficult-to-treat infections in humans and hospital-acquired-infections. This review discusses the epidemiology, microbiology, diagnosis, and treatment of MDRSE infection and identifies knowledge gaps. By using the search term “pan resistant
Staphylococcus epidermidis” OR “multi-drug resistant
Staphylococcus epidermidis” OR “multidrug-resistant lineages of
Staphylococcus epidermidis”, a total of 64 records have been identified from various previously published studies. The proportion of methicillin resistance in
S. epidermidis has been reported to be as high as 92%. Several studies across the world have aimed to detect the main phylogenetic lineages and antibiotically resistant genes through culture, mass spectrometry, and genomic analysis. Molecular biology tools are now available for the identification of
S. epidermidis and its drug resistance mechanisms, especially in blood cultures. However, understanding the distinction between a simple colonization and a bloodstream infection (BSI) caused by
S. epidermidis is still a challenge for clinicians. Some important parameters to keep in mind are the number of positive samples, the symptoms and signs of the patient, the comorbidities of the patient, the presence of central venous catheter (CVC) or other medical device, and the resistance phenotype of the organism. The agent of choice for empiric parenteral therapy is vancomycin. Other treatment options, depending on different clinical settings, may include teicoplanin, daptomycin, oxazolidinones, long-acting lipoglycopeptides, and ceftaroline. For patients with
S. epidermidis infections associated with the presence of an indwelling device, assessment regarding whether the device warrants removal is an important component of management. This study provides an overview of the MDRSE infection. Further studies are needed to explore and establish the most correct form of management of this infection.
Full article
►▼
Show Figures
Open AccessArticle
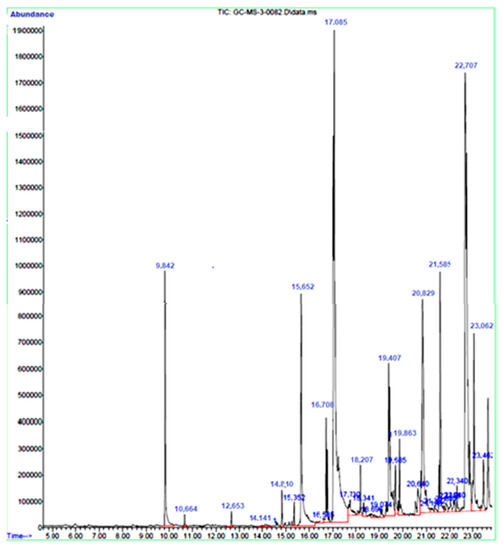
Antimicrobial Activities and Biofilm Inhibition Properties of Trigonella foenumgraecum Methanol Extracts against Multidrug-Resistant Staphylococcus aureus and Escherichia coli
by
Rawaf Alenazy
Cited by 3 | Viewed by 2352
Abstract
Multidrug-resistant bacteria are becoming the leading cause of death globally due to their resistance to many currently used antibiotics. Bacteria naturally have intrinsic resistance or acquired resistance to certain commonly used antibiotics. Therefore, searching for novel compounds has become necessary.
Trigonella foenumgraecum extract
[...] Read more.
Multidrug-resistant bacteria are becoming the leading cause of death globally due to their resistance to many currently used antibiotics. Bacteria naturally have intrinsic resistance or acquired resistance to certain commonly used antibiotics. Therefore, searching for novel compounds has become necessary.
Trigonella foenumgraecum extract was evaluated for antimicrobial and antibiofilm activities against multidrug-resistant bacteria
Staphylococcus aureus and
Escherichia coli. The minimum inhibitory concentration and minimum bactericidal concentration of the extract were also determined. Moreover, gas chromatography-mass spectrometry (GC-MS) analysis was used to identify the phytochemical components present in the extract. GC-MS analysis revealed that
T. foenumgraecum extract contains major compounds such as Phenol, 2-methoxy-3-(2-propenyl)-, n-Hexadecanoic acid, and 9,12,15-Octadecatrienoic acid. Both bacterial strains showed resistance to some of the antibiotics tested.
T. foenumgraecum showed inhibitory activity against the tested bacterial strains with a MIC of 500 µg/mL and MBC of 1000 µg/mL. The methanol extract decreased the biofilm activity of both
E. coli and
S. aureus below the sub-minimum inhibitory concentration. The extract showed antibacterial and antibiofilm activity against the tested bacterial pathogens.
Full article
►▼
Show Figures
Open AccessReview
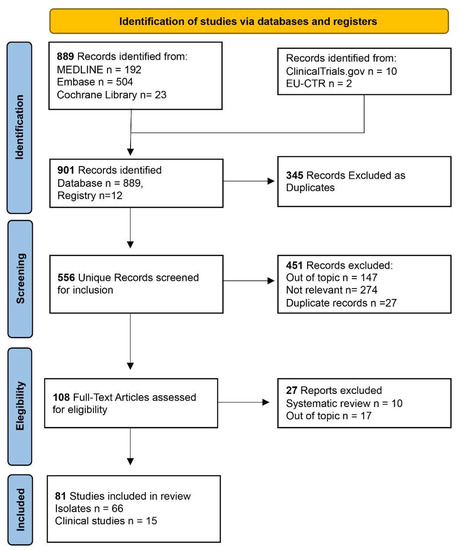
Plazomicin against Multidrug-Resistant Bacteria: A Scoping Review
by
Aniello Alfieri, Sveva Di Franco, Valerio Donatiello, Vincenzo Maffei, Ciro Fittipaldi, Marco Fiore, Francesco Coppolino, Pasquale Sansone, Maria Caterina Pace and Maria Beatrice Passavanti
Cited by 6 | Viewed by 1932
Abstract
Plazomicin is a next-generation semisynthetic aminoglycoside antibiotic that can be used to treat infections by multi-resistant bacteria. It is effective against many bacteria-producing carbapenemases or other specific hydrolases. This scoping review aims to define the role acquired by plazomicin from its approval by
[...] Read more.
Plazomicin is a next-generation semisynthetic aminoglycoside antibiotic that can be used to treat infections by multi-resistant bacteria. It is effective against many bacteria-producing carbapenemases or other specific hydrolases. This scoping review aims to define the role acquired by plazomicin from its approval by the FDA (US Food and Drug Administration) in 2018 to the present day. Furthermore, we aim to provide a base for a future meta-analysis. This project was conducted following the recommendations presented in the PRISMA extension for scoping reviews and the JBI Manual for Evidence Synthesis. Among 901 potentially engaging citations, 345 duplicates were removed, and only 81 articles were selected for the analysis. According to the data analysis, plazomicin has been used to treat urinary tract infections, bloodstream infections, and ventilation-associated pneumonia. The pathogens killed included multi-resistant
E. coli,
K. pneumoniae,
A. baumannii,
P. aeruginosa, and
S. aureus. Plazomicin can be a manageable, valid non-beta-lactam alternative for treating multi-resistant bacteria infections.
Full article
►▼
Show Figures
Open AccessPerspective
Multidrug Resistence Prevalence in COVID Area
by
Caterina Aurilio, Pasquale Sansone, Antonella Paladini, Manlio Barbarisi, Francesco Coppolino, Vincenzo Pota and Maria Caterina Pace
Cited by 12 | Viewed by 2561
Abstract
Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, is often complicated by severe acute respiratory syndrome. The new coronavirus outbreak started in China in December 2019 and rapidly spread around the world. The high diffusibility of the virus was the reason for the outbreak
[...] Read more.
Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, is often complicated by severe acute respiratory syndrome. The new coronavirus outbreak started in China in December 2019 and rapidly spread around the world. The high diffusibility of the virus was the reason for the outbreak of the pandemic viral disease, reaching more than 100 million infected people globally by the first three months of 2021. In the various treatments used up to now, the use of antimicrobial drugs for the management, especially of bacterial co-infections, is very frequent in patients admitted to intensive care. In addition, critically ill patients with SARS-CoV-2 infection are subjected to prolonged mechanical ventilation and other therapeutic procedures often responsible for developing hospital co-infections due to multidrug-resistant bacteria. Co-infections contribute to the increase in the morbidity–mortality of viral respiratory infections. We performed this study to review the recent articles published on the antibiotic bacterial resistance and viruses to predict risk factors of coronavirus disease 2019 and to assess the multidrug resistance in patients hospitalized in the COVID-19 area.
Full article
Open AccessReview
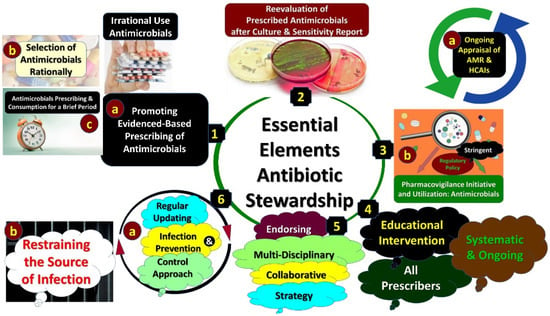
Strategies to Improve Antimicrobial Utilization with a Special Focus on Developing Countries
by
Brian Godman, Abiodun Egwuenu, Mainul Haque, Oliver Ombeva Malande, Natalie Schellack, Santosh Kumar, Zikria Saleem, Jacqueline Sneddon, Iris Hoxha, Salequl Islam, Julius Mwita, Renata Cristina Rezende Macedo do Nascimento, Isabella Piassi Dias Godói, Loveline Lum Niba, Adefolarin A. Amu, Joseph Acolatse, Robert Incoom, Israel Abebrese Sefah, Sylvia Opanga, Amanj Kurdi, Ibrahim Chikowe, Felix Khuluza, Dan Kibuule, Olayinka O. Ogunleye, Adesola Olalekan, Vanda Markovic-Pekovic, Johanna C. Meyer, Abubakr Alfadl, Thuy Nguyen Thi Phuong, Aubrey C. Kalungia, Stephen Campbell, Alice Pisana, Janney Wale and R. Andrew Seatonadd
Show full author list
remove
Hide full author list
| Viewed by 12812
Abstract
Antimicrobial resistance (AMR) is a high priority across countries as it increases morbidity, mortality and costs. Concerns with AMR have resulted in multiple initiatives internationally, nationally and regionally to enhance appropriate antibiotic utilization across sectors to reduce AMR, with the overuse of antibiotics
[...] Read more.
Antimicrobial resistance (AMR) is a high priority across countries as it increases morbidity, mortality and costs. Concerns with AMR have resulted in multiple initiatives internationally, nationally and regionally to enhance appropriate antibiotic utilization across sectors to reduce AMR, with the overuse of antibiotics exacerbated by the COVID-19 pandemic. Effectively tackling AMR is crucial for all countries. Principally a narrative review of ongoing activities across sectors was undertaken to improve antimicrobial use and address issues with vaccines including COVID-19. Point prevalence surveys have been successful in hospitals to identify areas for quality improvement programs, principally centering on antimicrobial stewardship programs. These include reducing prolonged antibiotic use to prevent surgical site infections. Multiple activities centering on education have been successful in reducing inappropriate prescribing and dispensing of antimicrobials in ambulatory care for essentially viral infections such as acute respiratory infections. It is imperative to develop new quality indicators for ambulatory care given current concerns, and instigate programs with clear public health messaging to reduce misinformation, essential for pandemics. Regular access to effective treatments is needed to reduce resistance to treatments for HIV, malaria and tuberculosis. Key stakeholder groups can instigate multiple initiatives to reduce AMR. These need to be followed up.
Full article
►▼
Show Figures
Open AccessReview
Emerging Treatment Options for Multi-Drug-Resistant Bacterial Infections
by
Roberto Giurazza, Maria Civita Mazza, Roberto Andini, Pasquale Sansone, Maria Caterina Pace and Emanuele Durante-Mangoni
Cited by 31 | Viewed by 6022
Abstract
Antimicrobial resistance (AMR) remains one of the top public health issues of global concern. Among the most important strategies for AMR control there is the correct and appropriate use of antibiotics, including those available for the treatment of AMR pathogens. In this article,
[...] Read more.
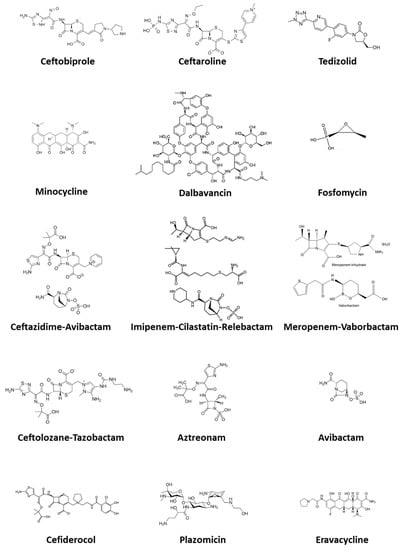
Antimicrobial resistance (AMR) remains one of the top public health issues of global concern. Among the most important strategies for AMR control there is the correct and appropriate use of antibiotics, including those available for the treatment of AMR pathogens. In this article, after briefly reviewing the most important and clinically relevant multi-drug-resistant bacteria and their main resistance mechanisms, we describe the emerging antimicrobial options for both MDR Gram-positive cocci and Gram-negative bacilli, including recently marketed agents, molecules just approved or under evaluation and rediscovered older antibiotics that have regained importance due to their antimicrobial spectrum. Specifically, emerging options for Gram-positive cocci we reviewed include ceftaroline, ceftobiprole, tedizolid, dalbavancin, and fosfomycin. Emerging treatment options for Gram-negative bacilli we considered comprise ceftolozane-tazobactam, ceftazidime-avibactam, meropenem-vaborbactam, imipenem-relebactam, aztreonam-avibactam, minocycline, fosfomycin, eravacycline, plazomicin, and cefiderocol. An exciting scenario is opening today with the long awaited growing availability of novel molecules for the treatment of AMR bacteria. Knowledge of mechanisms of action and resistance patterns allows physicians to increasingly drive antimicrobial treatment towards a precision medicine approach. Strict adherence to antimicrobial stewardship practices will allow us to preserve the emerging antimicrobials for our future.
Full article
►▼
Show Figures
Open AccessArticle
Inappropriateness of Antibiotic Prescribing in Medical, Surgical and Intensive Care Units: Results of a Multicentre Observational Study
by
Margherita Macera, Federica Calò, Lorenzo Onorato, Giovanni Di Caprio, Caterina Monari, Antonio Russo, Anna Galdieri, Antonio Giordano, Patrizia Cuccaro and Nicola Coppola
Cited by 6 | Viewed by 1808
Abstract
The objectives of the present study were to provide a snapshot analysis of antibiotic appropriateness in two hospitals in Southern Italy in three specific areas, surgical, medical and intensive care, and to evaluate the risk factors associated with inappropriateness in antimicrobial prescriptions. We
[...] Read more.
The objectives of the present study were to provide a snapshot analysis of antibiotic appropriateness in two hospitals in Southern Italy in three specific areas, surgical, medical and intensive care, and to evaluate the risk factors associated with inappropriateness in antimicrobial prescriptions. We conducted a multicentre observational study in two hospitals in the Campania region. We collected data of all patients admitted on the day of evaluation to antibiotic therapy or prophylaxis through a case report form. The primary outcome was to assess the inappropriateness of antibiotic prescribing, related to the spectrum, dose, route of administration and duration of treatment—in particular, to assess whether there was a difference in the adequacy of the prescriptive practice in the medical, surgical and intensive sectors. Prescriptive inappropriateness was more frequently observed in surgical units (79.8% of the 104 antimicrobial prescriptions) than in medical units (53.8% of the 65 prescriptions,
p = 0.0003) or in intensive care units (64.1% of the 39 prescriptions,
p = 0.052). The reasons for the inappropriate antimicrobial prescriptions were similar in the three areas evaluated: antimicrobial unnecessary and antimicrobial not recommended were the most frequent reasons for inappropriateness. Not participating in an antimicrobial stewardship program (ASP) was identified as a factor associated with inappropriate antimicrobial prescriptions in medical and surgical units, but not in Intensive Care Units (ICUs). ASPs may enhance the appropriateness of antimicrobial prescriptions especially in medical and surgical units. In ICUs, specific programs able to limit empirical therapies and encourage the collection of microbiological samples may be useful to set up targeted therapies and to design antimicrobial protocols.
Full article
Open AccessReview
Ceftolozane/Tazobactam for Resistant Drugs Pseudomonas aeruginosa Respiratory Infections: A Systematic Literature Review of the Real-World Evidence
by
Luca Gregorio Giaccari, Maria Caterina Pace, Maria Beatrice Passavanti, Francesca Gargano, Caterina Aurilio and Pasquale Sansone
Cited by 8 | Viewed by 2542
Abstract
Background: Ceftolozane/tazobactam (C/T) is a β-lactam/β-lactamase inhibitor combination that mainly targets Gram-negative bacteria. The current international guidelines recommend including C/T treatment in the empirical therapy for hospital-acquired pneumonia (HAP) and ventilator-associated pneumonia (VAP).
Pseudomonas aeruginosa (PA) is one of the most challenging Gram-negative
[...] Read more.
Background: Ceftolozane/tazobactam (C/T) is a β-lactam/β-lactamase inhibitor combination that mainly targets Gram-negative bacteria. The current international guidelines recommend including C/T treatment in the empirical therapy for hospital-acquired pneumonia (HAP) and ventilator-associated pneumonia (VAP).
Pseudomonas aeruginosa (PA) is one of the most challenging Gram-negative bacteria. We conducted a systematic review of all cases reported in the literature to summarize the existing evidence. Methods: The main electronic databases were screened to identify case reports of patients with drug-resistant PA respiratory infections treated with C/T. Results: A total of 22 publications were included for a total of 84 infective episodes. The clinical success rate was 72.6% across a wide range of comorbidities. The 45.8% of patients treated with C/T presented colonization by PA. C/T was well tolerated. Only six patients presented adverse events, but none had to stop treatment. The most common therapeutic regimens were 1.5 g every 8 h and 3 g every 8 h. Conclusion: C/T may be a valid therapeutic option to treat multidrug-resistant (MDR), extensively drug-resistant (XDR), pandrug-resistant (PDR), and carbapenem-resistant (CR) PA infections. However, further data are necessary to define the optimal treatment dosage and duration.
Full article
►▼
Show Figures
Open AccessArticle
Shewanella algae and Morganella morganii Coinfection in Cobra-Bite Wounds: A Genomic Analysis
by
Wei-Hsuan Huang, Chin-Chuan Kao, Yan-Chiao Mao, Chih-Sheng Lai, Kuo-Lung Lai, Chung-Hsu Lai, Chien-Hao Tseng, Yao-Ting Huang and Po-Yu Liu
Cited by 3 | Viewed by 2024
Abstract
Naja atra bites cause severe soft tissue injury and are prone to wound infections. The pathogens of
Naja atra bite-wound infections are highly variable in different geographical regions. Here, we report the first coinfection with
Shewanella algae and
Morganella morganii from a
Naja
[...] Read more.
Naja atra bites cause severe soft tissue injury and are prone to wound infections. The pathogens of
Naja atra bite-wound infections are highly variable in different geographical regions. Here, we report the first coinfection with
Shewanella algae and
Morganella morganii from a
Naja atra bite wound with resistome analysis using whole genome sequencing.
Full article
►▼
Show Figures
Open AccessArticle
Postoperative Complications Are Associated with Long-Term Changes in the Gut Microbiota Following Colorectal Cancer Surgery
by
Felix C. F. Schmitt, Martin Schneider, William Mathejczyk, Markus A. Weigand, Jane C. Figueiredo, Christopher I. Li, David Shibata, Erin M. Siegel, Adetunji T. Toriola, Cornelia M. Ulrich, Alexis B. Ulrich, Sébastien Boutin and Biljana Gigic
Cited by 9 | Viewed by 2428
Abstract
Changes in the gut microbiome have already been associated with postoperative complications in major abdominal surgery. However, it is still unclear whether these changes are transient or a long-lasting effect. Therefore, the aim of this prospective clinical pilot study was to examine long-term
[...] Read more.
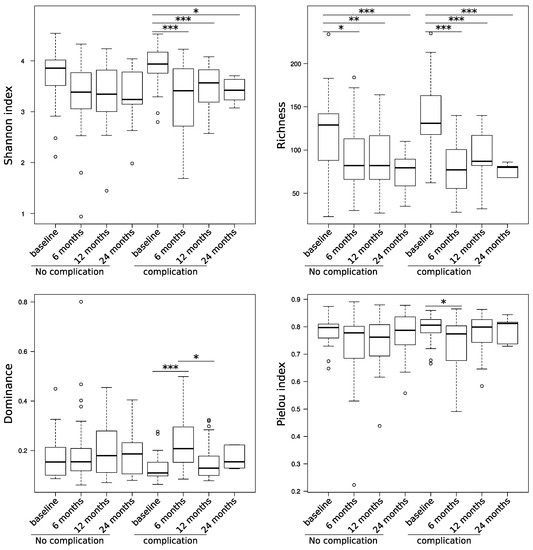
Changes in the gut microbiome have already been associated with postoperative complications in major abdominal surgery. However, it is still unclear whether these changes are transient or a long-lasting effect. Therefore, the aim of this prospective clinical pilot study was to examine long-term changes in the gut microbiota and to correlate these changes with the clinical course of the patient. Methods: In total, stool samples of 62 newly diagnosed colorectal cancer patients undergoing primary tumor resection were analyzed by 16S-rDNA next-generation sequencing. Stool samples were collected preoperatively in order to determine the gut microbiome at baseline as well as at 6, 12, and 24 months thereafter to observe longitudinal changes. Postoperatively, the study patients were separated into two groups—patients who suffered from postoperative complications (
n = 30) and those without complication (
n = 32). Patients with postoperative complications showed a significantly stronger reduction in the alpha diversity starting 6 months after operation, which does not resolve, even after 24 months. The structure of the microbiome was also significantly altered from baseline at six-month follow-up in patients with complications (
p = 0.006). This was associated with a long-lasting decrease of a large number of species in the gut microbiota indicating an impact in the commensal microbiota and a long-lasting increase of
Fusobacterium ulcerans. The microbial composition of the gut microbiome shows significant changes in patients with postoperative complications up to 24 months after surgery.
Full article
►▼
Show Figures
Open AccessReview
Implications of COVID-19 Pandemic on the Emergence of Antimicrobial Resistance: Adjusting the Response to Future Outbreaks
by
Doris Rusic, Marino Vilovic, Josipa Bukic, Dario Leskur, Ana Seselja Perisin, Marko Kumric, Dinko Martinovic, Ana Petric, Darko Modun and Josko Bozic
Cited by 33 | Viewed by 5624
Abstract
The net effect of the coronavirus disease 2019 (COVID-19) pandemic and the response to it on the emergence of antimicrobial resistance is yet unknown. Positive impacts on the spread of multiresistant pathogens and infections in general may be observed with the implementation of
[...] Read more.
The net effect of the coronavirus disease 2019 (COVID-19) pandemic and the response to it on the emergence of antimicrobial resistance is yet unknown. Positive impacts on the spread of multiresistant pathogens and infections in general may be observed with the implementation of general preventative measures for the spread of infectious disease such as social distancing, reduced travel and increased personal hygiene. This pandemic has accelerated the development of novel technologies, such as mRNA vaccines, that may be used to fight other diseases. These should be capitalized upon to manage the ongoing antimicrobial resistance pandemic in the background. However, it is likely that the COVID-19 pandemic is fueling the emergence of antimicrobial resistance due to high rates of inappropriate antimicrobial prescribing, the high use of biocides and the interruption of treatment for other conditions. Clinical uncertainty driven by the lack of effective diagnostics and practice of telemedicine may have driven the inappropriate use of antimicrobials. As pathogens know no borders, increased focus is needed for infectious diseases still threatening low- and middle-income countries such as tuberculosis. Stewardship measures for future outbreaks should stress the importance of social distancing and hand washing but discourage the overuse of disinfectants and antimicrobials that are not proven effective.
Full article
Open AccessArticle
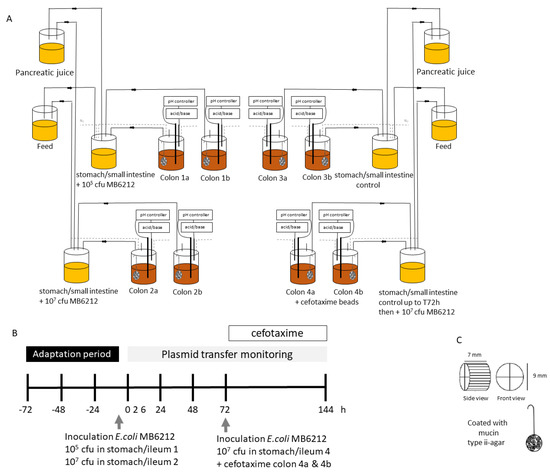
Transfer of Antibiotic Resistance Plasmid from Commensal E. coli towards Human Intestinal Microbiota in the M-SHIME: Effect of E. coli dosis, Human Individual and Antibiotic Use
by
Ellen Lambrecht, Els Van Coillie, Nico Boon, Marc Heyndrickx and Tom Van de Wiele
Cited by 5 | Viewed by 2515
Abstract
Along with (in)direct contact with animals and a contaminated environment, humans are exposed to antibiotic resistant bacteria by consumption of food. The implications of ingesting antibiotic resistant commensal bacteria are unknown, as dose-response data on resistance transfer and spreading in our gut is
[...] Read more.
Along with (in)direct contact with animals and a contaminated environment, humans are exposed to antibiotic resistant bacteria by consumption of food. The implications of ingesting antibiotic resistant commensal bacteria are unknown, as dose-response data on resistance transfer and spreading in our gut is lacking. In this study, transfer of a resistance plasmid (IncF), harbouring several antibiotic resistance genes, from a commensal
E. coli strain towards human intestinal microbiota was assessed using a Mucosal Simulator of the Human Intestinal Ecosystem (M-SHIME). More specifically, the effect of the initial
E. coli plasmiddonor concentration (10
5 and 10
7 CFU/meal), antibiotic treatment (cefotaxime) and human individual (n = 6) on plasmid transfer towards lumen coliforms and anaerobes was determined. Transfer of the resistance plasmid to luminal coliforms and anaerobes was observed shortly after the donor strain arrived in the colon and was independent of the ingested dose. Transfer occurred in all six simulated colons and despite their unique microbial community composition, no differences could be detected in antibiotic resistance transfer rates between the simulated human colons. After 72 h, resistant coliform transconjugants levels ranged from 7.6 × 10
4 to 7.9 × 10
6 CFU
cefotaxime resistant/mL colon lumen. Presence of the resistance plasmid was confirmed and quantified by PCR and qPCR. Cefotaxime treatment led to a significant reduction (85%) in resistant coliforms, however no significant effect on the total number of cultivable coliforms and anaerobes was observed.
Full article
►▼
Show Figures
Open AccessArticle
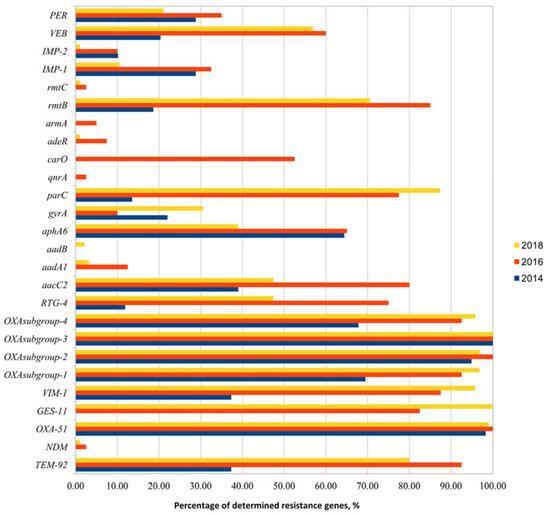
Multidrug-Resistant Acinetobacter baumannii Genetic Characterization and Spread in Lithuania in 2014, 2016, and 2018
by
Tatjana Kirtikliene, Aistė Mierauskaitė, Ilona Razmienė and Nomeda Kuisiene
Cited by 2 | Viewed by 2800
Abstract
Bacterial resistance to antimicrobial agents plays an important role in the treatment of bacterial infections in healthcare institutions. The spread of multidrug-resistant bacteria can occur during inter- and intra-hospital transmissions among patients and hospital personnel. For this reason, more studies must be conducted
[...] Read more.
Bacterial resistance to antimicrobial agents plays an important role in the treatment of bacterial infections in healthcare institutions. The spread of multidrug-resistant bacteria can occur during inter- and intra-hospital transmissions among patients and hospital personnel. For this reason, more studies must be conducted to understand how resistance occurs in bacteria and how it moves between hospitals by comparing data from different years and looking out for any patterns that might emerge. Multidrug-resistant (MDR)
Acinetobacter spp. was studied at 14 healthcare institutions in Lithuania during 2014, 2016, and 2018 using samples from human bloodstream infections. In total, 194 isolates were collected and identified using MALDI-TOF and VITEK2 analyzers as
Acinetobacter baumannii group bacteria. After that, the isolates were analyzed for the presence of different resistance genes (20 genes were analyzed) and characterized by using the Rep-PCR and MLVA (multiple-locus variable-number tandem repeat analysis) genotyping methods. The results of the study showed the relatedness of the different
Acinetobacter spp. isolates and a possible circulation of resistance genes or profiles during the different years of the study. This study provides essential information, such as variability and diversity of resistance genes, genetic profiling, and clustering of isolates, to better understand the antimicrobial resistance patterns of
Acinetobacter spp. These results can be used to strengthen the control of multidrug-resistant infections in healthcare institutions and to prevent potential outbreaks of this pathogen in the future.
Full article
►▼
Show Figures
Open AccessArticle
Detection of Carbapenem-Resistant Enterobacterales in Simulated Blood Culture in 15 Minutes
by
Daria Baer, Maya Azrad, Nora Saleh and Avi Peretz
Cited by 5 | Viewed by 2155
Abstract
Bacteremia leading to sepsis and organ dysfunction is a life-threatening situation, leading to death of up to one fourth of the infected individuals around the world. One major challenge in the treatment of sepsis is the rising prevalence of antibiotic resistant bacteria, such
[...] Read more.
Bacteremia leading to sepsis and organ dysfunction is a life-threatening situation, leading to death of up to one fourth of the infected individuals around the world. One major challenge in the treatment of sepsis is the rising prevalence of antibiotic resistant bacteria, such as carbapenem-resistant Enterobacterales (CRE). In recent years, several molecular assays have been developed for the detection of CRE mechanisms, enabling rapid results reporting. We evaluated the performance of the NG-Test CARBA 5 (NG Biotech) kit in detection of CRE in simulated blood cultures. Carbapenemase-producing (CP) CRE isolates (
n = 38) and non-carbapenemase CRE (Non-CP) isolates (
n = 10), previously identified using the routine methods practiced at the clinical microbiology laboratory of the Baruch Padeh Medical Center, Israel, were used in this analysis. Variable concentrations of the bacterial isolates were added to a suspension composed of human blood and saline, simulating the composition of a blood culture. Samples were then transferred to an anaerobic blood culture bottle and later tested with the NG-Test CARBA 5 (NG Biotech) kit, that identifies the CRE mechanism within 15 min. The NG-Test CARBA 5 kit correctly identified 43 samples (89.5%). The sensitivity and specificity of the kits were 86.8% and 100%, respectively. In conclusion, the NG-Test CARBA 5 kit is a reliable and accessible tool for the rapid diagnosis of CRE bloodstream infections.
Full article
Open AccessCommunication
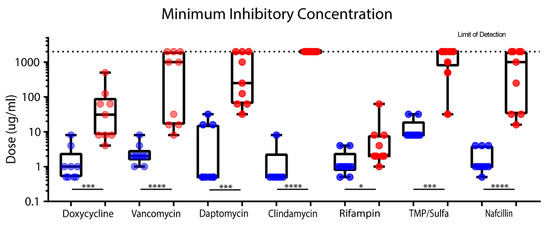
Staphylococcus epidermidis Biofilms Have a High Tolerance to Antibiotics in Periprosthetic Joint Infection
by
John A. Koch, Taylor M. Pust, Alex J. Cappellini, Jonathan B. Mandell, Dongzhu Ma, Neel B. Shah, Kimberly M. Brothers and Kenneth L. Urish
Cited by 18 | Viewed by 2906
Abstract
Both
Staphylococcus aureus and
Staphylococcus epidermidis are commonly associated with periprosthetic joint infections (PJIs). The treatment of PJI can be challenging because biofilms are assumed to have an increased intolerance to antibiotics. This makes the treatment of PJI challenging from a clinical perspective.
[...] Read more.
Both
Staphylococcus aureus and
Staphylococcus epidermidis are commonly associated with periprosthetic joint infections (PJIs). The treatment of PJI can be challenging because biofilms are assumed to have an increased intolerance to antibiotics. This makes the treatment of PJI challenging from a clinical perspective. Although
S. aureus has been previously demonstrated to have increased biofilm antibiotic tolerance, this has not been well established with
Staphylococcus epidermidis. A prospective registry of PJI
S. epidermidis isolates was developed. The efficacy of clinically relevant antibiotics was quantified against these isolates.
S. epidermidis planktonic minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) were collected using clinical laboratory standard index (CLSI) assays for eight antibiotics (doxycycline, vancomycin, daptomycin, clindamycin, rifampin, nafcillin, and trimethoprim/sulfamethoxazole). Mature biofilms were grown in vitro, after which minimum biofilm inhibitory concentration (MBIC) and minimum biofilm bactericidal concentration (MBBC) were quantified. Only rifampin and doxycycline had a measurable MBIC across all tested isolates. Based on MBBC, 64% of
S. epidermidis biofilms could be eliminated by rifampin, whereas only 18% by doxycycline.
S. epidermidis biofilm was observed to have a high tolerance to antibiotics as compared to planktonic culture. Isolate biofilm antibiotic tolerance varied to a larger degree than was seen in planktonic cultures.
Full article
►▼
Show Figures
Planned Papers
The below list represents only planned manuscripts. Some of these
manuscripts have not been received by the Editorial Office yet. Papers
submitted to MDPI journals are subject to peer-review.
Title: Blood stream infections from MDR Bacteria
Authors: Maria Beatrice Passavanti
Affiliation: University of Campania "Luigi Vanvitelli
Title: MDR Pneumonia in Intensive Care
Authors: Pasquale Sansone
Affiliation: Department of Woman, Child and General and Specialized Surgery, University of Campania “Luigi Vanvitelli”, 81100 Naples, Italy
Title: Multidrug Resistance Mycosis
Authors: Antonella Paladini
Affiliation: Department of Life, Health and Environmental Sciences, University of L'Aquila, L'Aquila, Italy
Title: Adiuvant therapy for MDR Bacteria Sepsis
Authors: Vincenzo Pota
Affiliation: Department of Woman, Child and General and Specialized Surgery, University of Campania “Luigi Vanvitelli”, 81100 Naples, Italy
Title: MDR Prevalence in COVID Area
Authors: Caterina Aurilio
Affiliation: Anesthesia and Intensive Care Unit, University of Campania Luigi Vanvitelli, 81100 Naples, Italy
Title: Emergent Antibiotics for MDR
Authors: Emanuele Durante Mangoni
Affiliation: Clinical & Experimental Medicine, Universita della Campania Vanvitelli, Italy
Title: The antimicrobial stewardship in management of MDR infections
Authors: Nicola Coppola
Affiliation: University of Campania, Italy
Title: Postoperative complications are associated with long-term changes in the gut microbiome following colorectal cancer surgery
Authors: Felix C.F. Schmitt1*, MD
Affiliation: Department of Anesthesiology, Heidelberg University Hospital, Heidelberg, Germany
Abstract: Background: Changes in the gut microbiome are already proven to be associated with postoperative complications in major abdominal surgery. However, it is still unclear whether these changes are transient or a long-lasting effect. Therefore, the aim of this prospective clinical pilot study was to examine long-term changes in the gut microbiome and to correlate these changes with the postoperative course of the patient. Methods: In total, stool samples of 62 newly-diagnosed colorectal cancer patients undergoing primary tumor resection were analysed by 16S-rDNA next-generation sequencing. Stool samples were collected preoperatively in order to determine the baseline gut microbiome as well as 6, 12, and 24 months thereafter to observe longitudinal changes. Results: Postoperatively, the participating patients were classified into two groups - patients who suffered from postoperative complications (n=30) and those without complication (n=32). Patients with postoperative complications showed a significantly stronger reduction in the alpha diversity which does not resolve, even after 24 months. The structure of the microbiome was also significantly different between baseline and the 6 months follow-up in patients with complication (P=0.006). This was associated with a long-lasting decrease of a large number of species in the gut microbiota indicating an impact in the commensal microbiota and a long-lasting increase of Fusobacterium ulcerans. Conclusions: The microbial composition of the gut microbiome shows significant changes in patients with postoperative complications up to two years after surgery.
Title: Implications of COVID-19 pandemic on the emergence of antimicrobial resistance: adjusting the response to future outbreaks
Authors: Josko Bozic et al.
Affiliation: School of Medicine, University of Split, Split, Croatia