Recent Scientific Developments in Genetics, Genomics, and Breeding of Fruit Trees
Share This Topical Collection
Editors
 Dr. Junbei Ni
Dr. Junbei Ni
 Dr. Junbei Ni
Dr. Junbei Ni
E-Mail
Website
Collection Editor
College of Agriculture and Biotechnology, Zhejiang University, Hangzhou 310058, China
Interests: fruit germplasm resources and genetic breeding; fruit quality; secondary metabolism; light and hormone signal response; transcriptional and epigenetic regulation
Topical Collection Information
Dear Colleagues,
Fruit trees are cultivated all over the world due to their great economic value. In recent years, the increasing demand for high-quality fruit with high resistance to pests and diseases has tremendously promoted the development of fruit tree research. However, detailed functional analyses of fruit trees are far behind model plants, due to highly heterozygous and complex genomes and the barrier of plant genetic transformation and/or regeneration in some fruit tree species. Fortunately, the application of next-generation sequencing technologies coupled with rapid advances in bioinformatics has facilitated the development of fruit trees for genetic and genomic studies as well as molecular breeding over the last decade. Furthermore, the development of gene-editing technologies, such as CRISPR-Cas9, allows for their application in the study of fruit tree genetics and breeding.
The proposed Topical Collection entitled “Recent Scientific Developments in Genetics, Genomics, and Breeding of Fruit Trees” aims to present advances in gene mining, genetic mechanisms, omics analyses (e.g., genomics, transcriptomics and proteomics), and the application of gene-editing technologies and molecular breeding of fruit trees. We look forward to receiving your manuscripts (including reviews and research articles) and are eager to share your newest findings.
Dr. Minjie Qian
Dr. Aidi Zhang
Dr. Junbei Ni
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Horticulturae is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2200 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- fruit trees
- molecular breeding
- genotyping and phenotyping
- bioinformatics
- comparative genomics
- QTL mapping
- genome-wide association studies
- transcriptome analysis
- gene editing
- gene function
Published Papers (5 papers)
Open AccessArticle
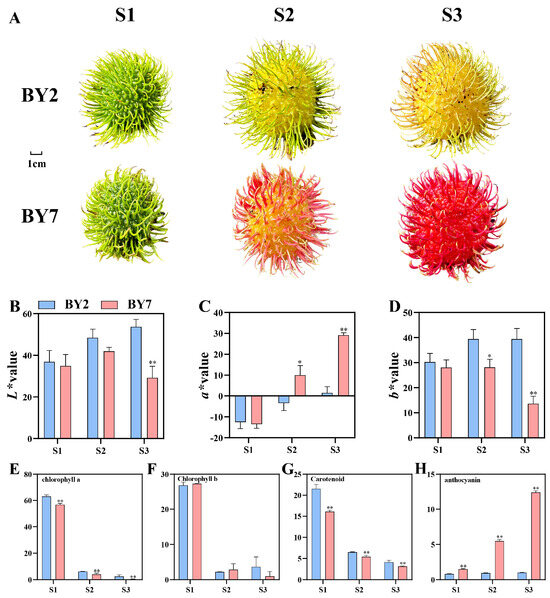
Comparative Metabolomic and Transcriptomic Analysis Reveals That Variations in Flavonoids Determine the Colors of Different Rambutan Cultivars
by
Jiaqi Wang, Wencan Zhu, Chengkun Yang, Maofu Li, Shun Feng, Lizhu Tang, Chengchao Yang and Zhifu Cui
Viewed by 615
Abstract
Rambutan is a tropical tree and its fruit has several favorable characteristics. To understand how the color of the rambutan fruit peel develops, the transcriptome, flavonoid metabolome, and carotenoid metabolome data of two rambutan cultivars, ‘BY2’ and ‘BY7’, which show yellow and red
[...] Read more.
Rambutan is a tropical tree and its fruit has several favorable characteristics. To understand how the color of the rambutan fruit peel develops, the transcriptome, flavonoid metabolome, and carotenoid metabolome data of two rambutan cultivars, ‘BY2’ and ‘BY7’, which show yellow and red peels at maturity, respectively, were comprehensively analyzed at three developmental stages. We identified 26 carotenoid components and 53 flavonoid components in these cultivars. Anthocyanins were the main component contributing to the red color of ‘BY7’ after reaching ripeness. The carotenoid content decreased sharply as the fruit matured. Hence, we speculated that flavonols were the main contributors to the yellow color of the ‘BY2’ peel. In total, 6805 differentially expressed genes were screened by transcriptome analysis; the majority of them were enriched in metabolic pathways and the biosynthesis of secondary metabolites. Weighted gene co-expression network analysis results revealed that in addition to MYB and bHLH, ERF, WRKY, MYB-related, and C3H were the main potential transcription factors regulating the color of the rambutan peel. In addition, we also identified 12 structural genes associated with flavonoid biosynthesis. The research findings shed light on the molecular mechanisms of color acquisition in rambutan fruit peels, laying the foundation for the quality control of rambutan and the cultivation of differently colored cultivars of rambutan.
Full article
►▼
Show Figures
Open AccessArticle
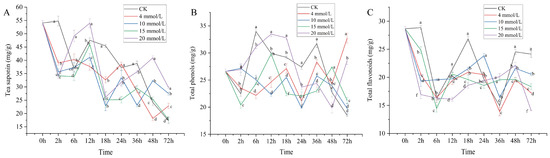
Physiological and Transcriptional Analysis Provides Insights into Tea Saponin Biosynthesis and Regulation in Response to SA in Camellia vietnamensis Huang
by
Yang Li, Heqin Yan, Muhammad Zeeshan Ul Haq, Ya Liu, Yougen Wu, Jing Yu and Pengguo Xia
Viewed by 817
Abstract
Camellia vietnamensis Huang is an important and famous woody oil crop with high economic value in China because of its high-quality, edible, and medicinal oil. As one of its major active components, tea saponin (triterpenoid saponin) has shown anticancer, antioxidant, bacteriostatic, and other
[...] Read more.
Camellia vietnamensis Huang is an important and famous woody oil crop with high economic value in China because of its high-quality, edible, and medicinal oil. As one of its major active components, tea saponin (triterpenoid saponin) has shown anticancer, antioxidant, bacteriostatic, and other pharmacological activities. In this study,
C. vietnamensis was used as an experimental material to determine the tea saponin content and physiological activity indicators after salicylic acid (SA) treatment and to analyze the differential expression genes of key metabolic pathways in response to SA by combining transcriptome data. The results showed that SA treatment increased the content of tea saponin and total phenols in leaves; effectively promoted the activity of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX); and decreased the content of malondialdehyde (MDA). A total of 60,038 genes, including 5871 new genes, were obtained by the RNA-seq. There were 6609 significantly differential expression genes mainly enriched in pathways such as sesquiterpenoid and triterpenoid biosynthesis, terpenoid backbone biosynthesis, diterpenoid biosynthesis, and flavonoid biosynthesis. The SA-induced key structural genes (
SQS,
SQE,
bAS,
CYP450, and
UGT) and transcription factors related to the tea saponin biosynthetic pathway were screened by weighted gene co-expression network analysis (WGCNA). The results of this study could provide a theoretical basis and a new technical method to improve the content of tea saponin, with its excellent anticancer activity, in
C. vietnamensis.
Full article
►▼
Show Figures
Open AccessArticle
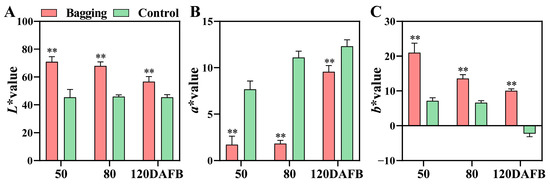
Transcription Regulation of Anthocyanins and Proanthocyanidins Accumulation by Bagging in ‘Ruby’ Red Mango: An RNA-seq Study
by
Wencan Zhu, Hongxia Wu, Chengkun Yang, Xiaowen Wang, Bin Shi, Bin Zheng, Xiaowei Ma, Minjie Qian, Aiping Gao and Kaibing Zhou
Viewed by 1009
Abstract
The biosynthesis of anthocyanins and proanthocyanidins (PAs), components of two main flavonoids in plants, is regulated by environmental factors such as light. We previously found that bagging significantly repressed the biosynthesis of anthocyanins in red ‘Ruby’ mango fruit peel, but induced the accumulation
[...] Read more.
The biosynthesis of anthocyanins and proanthocyanidins (PAs), components of two main flavonoids in plants, is regulated by environmental factors such as light. We previously found that bagging significantly repressed the biosynthesis of anthocyanins in red ‘Ruby’ mango fruit peel, but induced the accumulation of PAs. However, the molecular mechanism remains unclear. In the current study, transcriptome sequencing was used for screening the essential genes responsible for the opposite accumulation pattern of anthocyanins and PAs by bagging treatment. According to weighted gene co-expression network analysis (WGCNA), structural genes and transcription factors highly positively correlated to anthocyanins and PAs were identified. One flavanone 3-hydroxylase (
F3H) and seven structural genes, including one chalcone synthase (
CHS), one flavonoid 3’-hydroxylase (
F3’H), one anthocyanidin synthesis (
ANS), three leucoanthocyanidin reductase (
LARs), and one UDP glucose: flavonoid 3-O-glucosyltransferase (
UFGT), are crucial for anthocyanin and PA biosynthesis, respectively. In addition to MYB and bHLH, ERF, C2H2, HD-ZIP, and NAC are important transcription factors that participate in the regulation of anthocyanin and PA biosynthesis in ‘Ruby’ mango fruit peel by bagging treatment. Our results are helpful for revealing the transcription regulation mechanism of light-regulated mango anthocyanin and PA biosynthesis, developing new technologies for inducing flavonoid biosynthesis in mangos, and breeding mango cultivars containing high concentrations of flavonoids.
Full article
►▼
Show Figures
Open AccessArticle
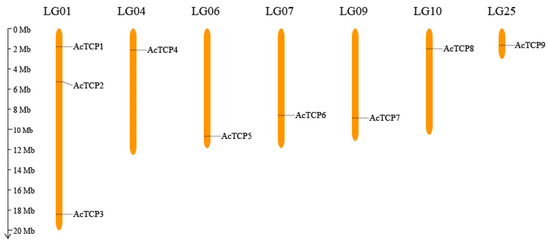
TCP Transcription Factors in Pineapple: Genome-Wide Characterization and Expression Profile Analysis during Flower and Fruit Development
by
Ziqiong Li, Yanwei Ouyang, Xiaolu Pan, Xiaohan Zhang, Lei Zhao, Can Wang, Rui Xu, Hongna Zhang and Yongzan Wei
Cited by 1 | Viewed by 958
Abstract
TEOSINTE-BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors contain specific a basic helix–loop–helix structure, which is a significant factor in the regulation of plant growth and development. TCP has been studied in several species, but no pineapple TCP has been reported to date. Whether they are involved
[...] Read more.
TEOSINTE-BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors contain specific a basic helix–loop–helix structure, which is a significant factor in the regulation of plant growth and development. TCP has been studied in several species, but no pineapple TCP has been reported to date. Whether they are involved in the development of the flower and fruit in the pineapple remains unclear. In this study, nine non-redundant pineapple TCPs (AcTCPs) were identified. Chromosomal localization, phylogenetics, gene structure, motifs, multiple-sequence alignment, and covariance on AcTCP family members were analyzed. Analysis of promoter
cis-acting elements illustrated that the
AcTCP gene may be mainly co-regulated by light signal and multiple hormone signals. Analysis of expression characteristics showed a significant increase in
AcTCP5 expression at 12 h after ethylene treatment, and significantly higher levels of
AcTCP8 and
AcTCP9 expression in the pistil than in other floral organs. Meanwhile, the
AcTCP4, AcTCP5, AcTCP6, AcTCP7, and
AcTCP9 expression levels were downregulated at later stages of fruit development. Transcription factors that may interact with TCP protein in the regulation of flower and fruit development are screened by the protein interaction prediction network, AcTCP5 interacts with AcSPL16, and AcTCP8 interacts with AcFT5 and AcFT6 proteins, verified by Y2H experiments. These findings provide a basis for further exploration of the molecular mechanisms and function of the
AcTCP gene in flower and fruit development.
Full article
►▼
Show Figures
Open AccessArticle
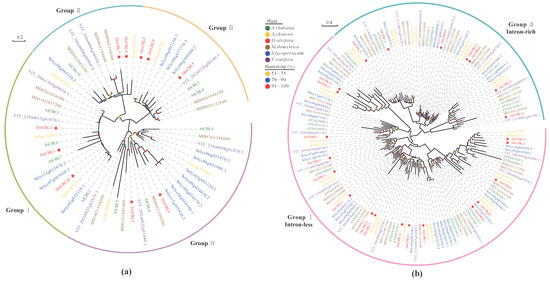
Genome–Wide Investigation of the CBL–CIPK Gene Family in Oil Persimmon: Evolution, Function and Expression Analysis during Development and Stress
by
Cuiyu Liu, Yanpeng Wang, Jin Yao, Xu Yang, Kaiyun Wu, Guoxin Teng, Bangchu Gong and Yang Xu
Cited by 1 | Viewed by 1308
Abstract
Ca
2+-sensors, calcineurin B-like proteins (CBLs), and calcineurin B-like protein-interacting protein kinases (CIPKs) form a CBL–CIPK complex to regulate signal transduction. This study aimed to reveal the characteristics of the CBL–CIPK gene family in oil persimmon (
Diospyros oleifera). Ten
DoCBL
[...] Read more.
Ca
2+-sensors, calcineurin B-like proteins (CBLs), and calcineurin B-like protein-interacting protein kinases (CIPKs) form a CBL–CIPK complex to regulate signal transduction. This study aimed to reveal the characteristics of the CBL–CIPK gene family in oil persimmon (
Diospyros oleifera). Ten
DoCBL and 23
DoCIPK genes were identified, and gene duplication among them was mainly attributed to segmental duplication. According to phylogenetic and structural analysis, DoCBLs were clustered into four groups with distinct motifs, namely myristoylation and palmytoylation sites in their N-terminus, and
DoCIPKs containing a NAF/FISL domain were clustered into intron-rich and intron-less groups. The expression patterns of
DoCBLs and
DoCIPKs were tissue- and time-specific in different tissues and at different stages of fruit development. Most
CBL–CIPK genes were upregulated under NaCl, drought, and Ca(NO
3)
2 stress using qRT-PCR analysis. DoCBL5 and DoCIPK05 were both located in the plasma membrane of cells using green fusion proteins (GFP) in tobacco leaves. DoCBL5 and DoCIPK05 might interact with AKT1, PP2C, and SNF to regulate the Ca
2+ signals, K
+, and ABA homeostasis in cells. In conclusion, these results suggested that the CBL–CIPK family genes might play important roles in oil persimmon growth and stress responses.
Full article
►▼
Show Figures