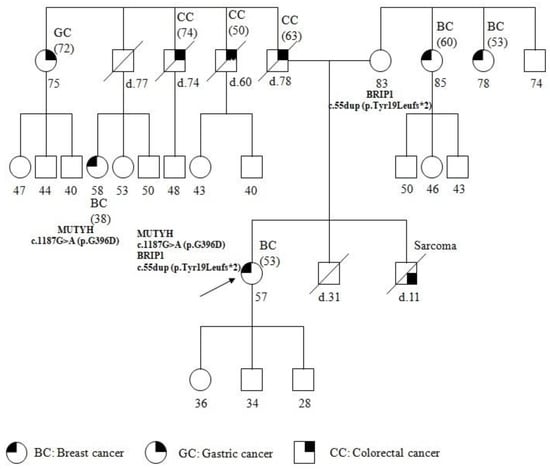
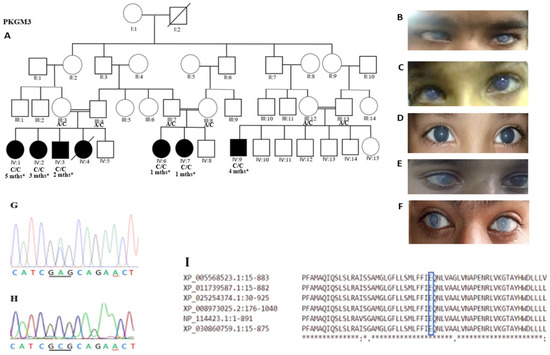
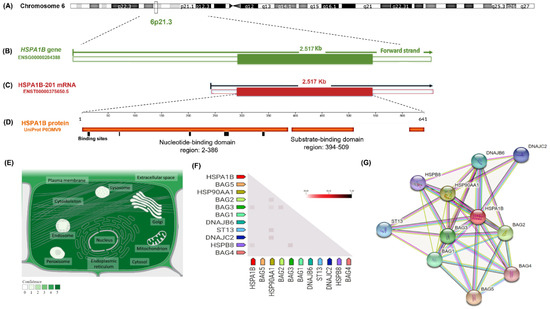
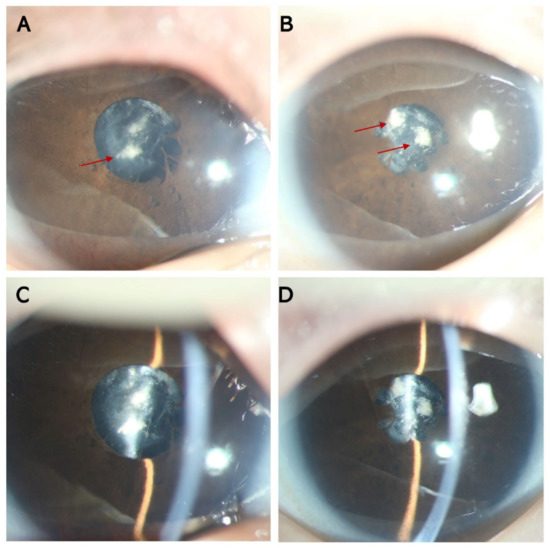
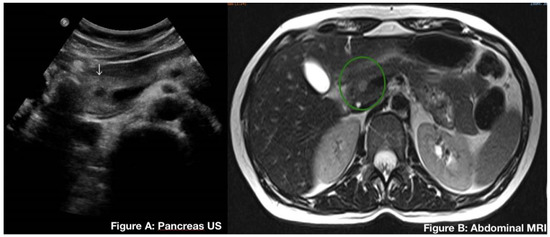
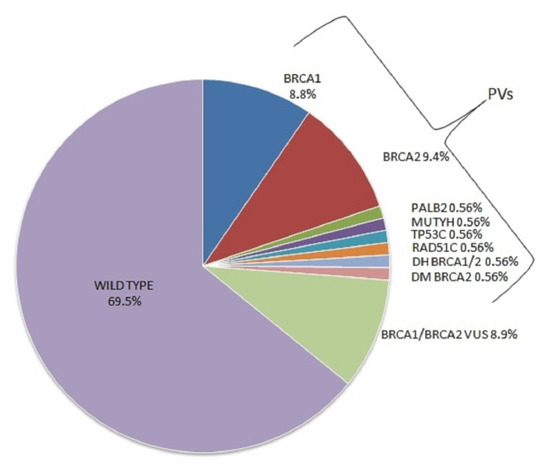
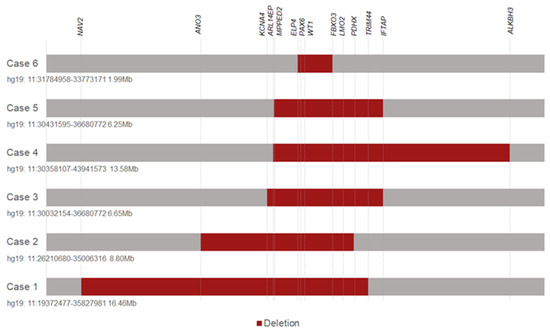
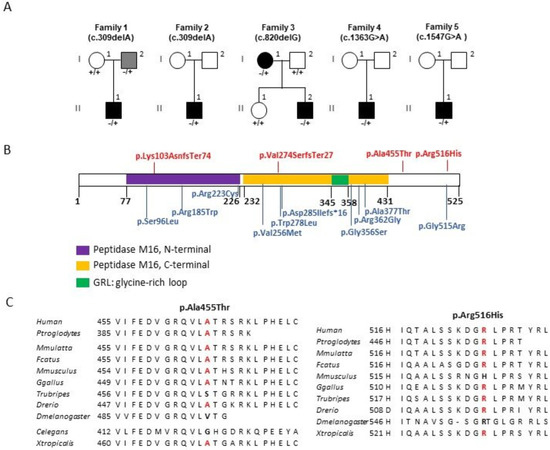
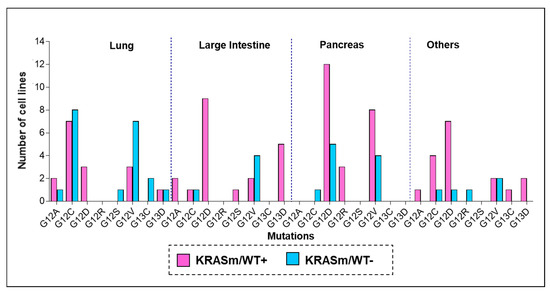
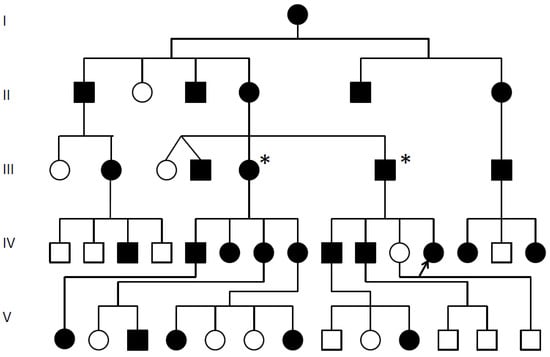
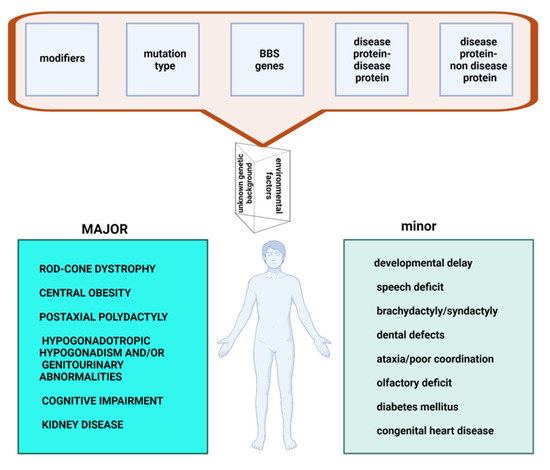
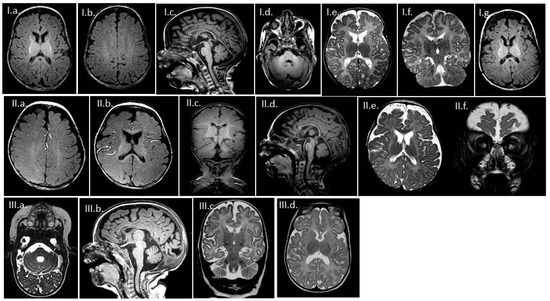
Genotype-Phenotype Study in Disease
A topical collection in Genes (ISSN 2073-4425). This collection belongs to the section "Human Genomics and Genetic Diseases".
Viewed by 102946Editors
Interests: molecular diagnosis; endocrine diseases; clinical pathology; tumor markers; metabolism disorders
Interests: hereditary cancer; molecular diagnosis; clinical pathology; genes related to hereditary cancer; DNA mutations and drug response; BRCA1, BRCA2 genes
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
The study of inter-individual variability is essential for precision medicine. It allows patients to be treated more specifically by identifying the most effective treatment. The study of polymorphic variants has become decisive in the understanding of mechanisms that underpin various multifactorial diseases (i.e., diabetes, obesity, cancer, and cardiovascular disease).
All diseases have different degrees of genetic heterogeneity, for which the severity of the disease varies from individual to individual. The same pathological phenotype could be determined by different mutations in different genes. In multifactorial diseases, however, genetic heterogeneity is much more widespread and frequent, so genes contribute to a different extent to the expression of phenotype.
Some genes do not cause disease but rather affect or cause particular aspects of the disease by behaving as modifying genes.
More recently it has been proposed that genetic variation may also explain some of the other features of clinical phenotype, such as disease duration.
Although the genotype–phenotype link is not always clear, it allows improved therapy, evaluation of individual drug response, and understanding of the adaptive response of organisms to environmental stimuli. In addition, in the economic and social spheres, the costs of the health system are reduced.
For these reasons, we believe it is important to prepare a Topical Collection with the title “Genotype-Phenotype Study in Disease”.
It would be a pleasure for us if you agreed to contribute your paper to this Topical Collection.
Prof. Dr. Michele Cioffi
Prof. Dr. Maria Teresa Vietri
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Genes is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Genotype-phenotype
- Genotype-guided therapy
- Multifactorial diseases
- Environmental stimuli
- Linking genotype and phenotype
- Personalized medicine
- Phenotypic heterogeneity
Planned Papers
The below list represents only planned manuscripts. Some of these manuscripts have not been received by the Editorial Office yet. Papers submitted to MDPI journals are subject to peer-review.
Author 1:
Tentative title: From HDLS to BANDDOS: fast-expanding genotype-phenotype association in CSF1R-caused disorders.
Author 2:
Tentative title: A novel mutation in Cu/Zn superoxide dismutase gene in ALS patients with flail leg variant