Epigenetic Control in Plants
A topical collection in Epigenomes (ISSN 2075-4655).
Viewed by 9767
Share This Topical Collection
Editor
 Dr. Vladimir Brukhin
Dr. Vladimir Brukhin
 Dr. Vladimir Brukhin
Dr. Vladimir Brukhin
E-Mail
Website
Collection Editor
1. Plant Genomics Group, ITMO University, Lomonosova, 9, 191002 Saint-Petersburg, Russia
2. Department of Plant Embryology & Reproductive Biology, Komarov Botanical Institute RAS, 2 Professor Popov Street, 197376 Saint-Petersburg, Russia
Interests: development; sexual and asexual reproduction; genomics
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Plants are sessile organisms with the capacity to respond to a varying environment throughout their lives. This capability is mediated through the moderation of gene expression without change to DNA sequence, a phenomenon known as epigenetics. Epigenetic mechanisms thereby mediate developmental progression of an organism and also the resilience to accommodate for change. Thus, epigenetic regulation in plants can be mediated in several ways, most notably mi RNA- and siRNA-based systems, histone modification and DNA methylation.
On a global scale methylation accumulates during somatic development, although external stimuli can cause either the methylation or demethylation of specific sites. About a third of plant genes are methylated at maturity but meiosis acts as a clearing house for methylation, with only a few methylated sites surviving through to the next generation. Atypical methylation can cause developmental or physiological anomalies.
The aim of this Topic Collection is to bring together a set of reviews and research articles on the role of epigenetic regulation in plants during sexual and asexual reproduction, development, and evolution.
Dr. Vladimir Brukhin
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Epigenomes is an international peer-reviewed open access quarterly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 1500 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- epigenetics
- methylation
- RNA interference
- chromatin remodeling
- plant reproduction
- development
Published Papers (3 papers)
Open AccessReview
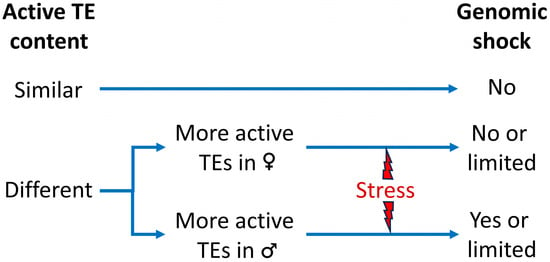
The Genomic Shock Hypothesis: Genetic and Epigenetic Alterations of Transposable Elements after Interspecific Hybridization in Plants
by
Carlos de Tomás and Carlos M. Vicient
Viewed by 1695
Abstract
Transposable elements (TEs) are major components of plant genomes with the ability to change their position in the genome or to create new copies of themselves in other positions in the genome. These can cause gene disruption and large-scale genomic alterations, including inversions,
[...] Read more.
Transposable elements (TEs) are major components of plant genomes with the ability to change their position in the genome or to create new copies of themselves in other positions in the genome. These can cause gene disruption and large-scale genomic alterations, including inversions, deletions, and duplications. Host organisms have evolved a set of mechanisms to suppress TE activity and counter the threat that they pose to genome integrity. These includes the epigenetic silencing of TEs mediated by a process of RNA-directed DNA methylation (RdDM). In most cases, the silencing machinery is very efficient for the vast majority of TEs. However, there are specific circumstances in which TEs can evade such silencing mechanisms, for example, a variety of biotic and abiotic stresses or in vitro culture. Hybridization is also proposed as an inductor of TE proliferation. In fact, the discoverer of the transposons, Barbara McClintock, first hypothesized that interspecific hybridization provides a “genomic shock” that inhibits the TE control mechanisms leading to the mobilization of TEs. However, the studies carried out on this topic have yielded diverse results, showing in some cases a total absence of mobilization or being limited to only some TE families. Here, we review the current knowledge about the impact of interspecific hybridization on TEs in plants and the possible implications of changes in the epigenetic mechanisms.
Full article
►▼
Show Figures
Open AccessReview
Epigenetic Modifications in Plant Development and Reproduction
by
Vladimir Brukhin and Emidio Albertini
Cited by 10 | Viewed by 4513
Abstract
Plants are exposed to highly fluctuating effects of light, temperature, weather conditions, and many other environmental factors throughout their life. As sessile organisms, unlike animals, they are unable to escape, hide, or even change their position. Therefore, the growth and development of plants
[...] Read more.
Plants are exposed to highly fluctuating effects of light, temperature, weather conditions, and many other environmental factors throughout their life. As sessile organisms, unlike animals, they are unable to escape, hide, or even change their position. Therefore, the growth and development of plants are largely determined by interaction with the external environment. The success of this interaction depends on the ability of the phenotype plasticity, which is largely determined by epigenetic regulation. In addition to how environmental factors can change the patterns of genes expression, epigenetic regulation determines how genetic expression changes during the differentiation of one cell type into another and how patterns of gene expression are passed from one cell to its descendants. Thus, one genome can generate many ‘epigenomes’. Epigenetic modifications acquire special significance during the formation of gametes and plant reproduction when epigenetic marks are eliminated during meiosis and early embryogenesis and later reappear. However, during asexual plant reproduction, when meiosis is absent or suspended, epigenetic modifications that have arisen in the parental sporophyte can be transmitted to the next clonal generation practically unchanged. In plants that reproduce sexually and asexually, epigenetic variability has different adaptive significance. In asexuals, epigenetic regulation is of particular importance for imparting plasticity to the phenotype when, apart from mutations, the genotype remains unchanged for many generations of individuals. Of particular interest is the question of the possibility of transferring acquired epigenetic memory to future generations and its potential role for natural selection and evolution. All these issues will be discussed to some extent in this review.
Full article
►▼
Show Figures
Open AccessEditorial
Epigenetic Control in Plants
by
Vladimir Brukhin
Viewed by 2340