Journal Description
Epigenomes
Epigenomes
is an international, peer-reviewed, open access journal on epigenetics and epigenomics, published quarterly online by MDPI. The Epigenetics Society is affiliated with Epigenomes and its members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, ESCI (Web of Science), PMC, PubMed, Embase, PubAg, CAPlus / SciFinder, and other databases.
- Journal Rank: CiteScore - Q2 (Biochemistry, Genetics and Molecular Biology (miscellaneous))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 15.3 days after submission; acceptance to publication is undertaken in 3.6 days (median values for papers published in this journal in the second half of 2023).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
2.5 (2022);
5-Year Impact Factor:
2.1 (2022)
Latest Articles
Deciphering the Diversity in Bacterial Transporters That Salvage Queuosine Precursors
Epigenomes 2024, 8(2), 16; https://doi.org/10.3390/epigenomes8020016 (registering DOI) - 25 Apr 2024
Abstract
Queuosine (Q) is a modification of the wobble base of tRNA harboring GUN anticodons with roles in decoding accuracy and efficiency. Its synthesis is complex with multiple enzymatic steps, and several pathway intermediates can be salvaged. The only two transporter families known to
[...] Read more.
Queuosine (Q) is a modification of the wobble base of tRNA harboring GUN anticodons with roles in decoding accuracy and efficiency. Its synthesis is complex with multiple enzymatic steps, and several pathway intermediates can be salvaged. The only two transporter families known to salvage Q precursors are QPTR/COG1738 and QrtT/QueT. Analyses of the distribution of known Q synthesis and salvage genes in human gut and oral microbiota genomes have suggested that more transporter families remain to be found and that Q precursor exchanges must occur within the structured microenvironments of the mammalian host. Using physical clustering and fusion-based association with Q salvage genes, candidate genes for missing transporters were identified and five were tested experimentally by complementation assays in Escherichia coli. Three genes encoding transporters from three different Pfam families, a ureide permease (PF07168) from Acidobacteriota bacterium, a hemolysin III family protein (PF03006) from Bifidobacterium breve, and a Major Facilitator Superfamily protein (PF07690) from Bartonella henselae, were found to allow the transport of both preQ0 and preQ1 in this heterologous system. This work suggests that many transporter families can evolve to transport Q precursors, reinforcing the concept of transporter plasticity.
Full article
(This article belongs to the Special Issue Epigenetic and Epitranscriptomic Determinants of Host-Microbe Interactions)
Open AccessArticle
TNFR1 Absence Is Not Crucial for Different Types of Cell Reaction to TNF: A Study of the TNFR1-Knockout Cell Model
by
Alina A. Alshevskaya, Julia A. Lopatnikova, Julia V. Zhukova, Olga Y. Perik-Zavodskaia, Saleh Alrhmoun, Irina A. Obleukhova, Anna K. Matveeva, Darya A. Savenkova, Ilnaz R. Imatdinov, Dmitry V. Yudkin and Sergey V. Sennikov
Epigenomes 2024, 8(2), 15; https://doi.org/10.3390/epigenomes8020015 - 03 Apr 2024
Abstract
►▼
Show Figures
Background: One of the mechanisms regulating the biological activity of tumor necrosis factor (TNF) in cells is the co-expression of TNFR1/TNFR2 receptors. A model with a differential level of receptor expression is required to evaluate the contribution of these mechanisms. Aim: The development
[...] Read more.
Background: One of the mechanisms regulating the biological activity of tumor necrosis factor (TNF) in cells is the co-expression of TNFR1/TNFR2 receptors. A model with a differential level of receptor expression is required to evaluate the contribution of these mechanisms. Aim: The development of a cellular model to compare the effects of TNF on cells depending on the presence of both receptors and TNFR2 alone. Methods: TNFR1 absence modifications of ZR-75/1 and K-562 cell lines were obtained by TNFR1 knockout. The presence of deletions was confirmed by Sanger sequencing, and the absence of cell membrane receptor expression was confirmed by flow cytometry. The dose-dependent effect of TNF on intact and knockout cells was comparatively evaluated by the effect on the cell cycle, the type of cell death, and the profile of expressed genes. Results: Knockout of TNFR1 resulted in a redistribution of TNFR2 receptors with an increased proportion of TNFR2+ cells in both lines and a multidirectional change in the density of expression in the lines (increased in K562 and decreased in ZR75/1). The presence of a large number of cells with high TNFR2 density in the absence of TNFR1 in the K562 cells was associated with greater sensitivity to TNF-stimulating doses and increased proliferation but did not result in a significant change in cell death parameters. A twofold increase in TNFR2+ cell distribution in this cell line at a reduced expression density in ZR75/1 cells was associated with a change in sensitivity to low cytokine concentrations in terms of proliferation; an overall increase in cell death, most pronounced at standard stimulating concentrations; and increased expression of the lymphocyte-activation gene groups, host–pathogen interaction, and innate immunity. Conclusions: The absence of TNFR1 leads to different variants of compensatory redistribution of TNFR2 in cellular models, which affects the type of cell response and the threshold level of sensitivity. The directionality of cytokine action modulation and sensitivity to TNF levels depends not only on the fraction of cells expressing TNFR2 but also on the density of expression.
Full article
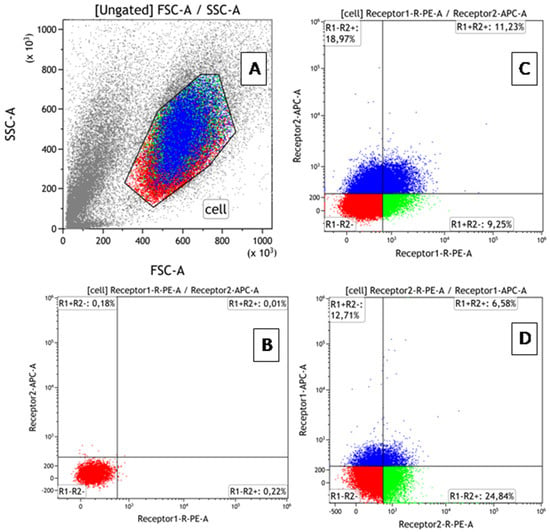
Figure 1
Open AccessArticle
Unveiling Gene Interactions in Alzheimer’s Disease by Integrating Genetic and Epigenetic Data with a Network-Based Approach
by
Keith L. Sanders, Astrid M. Manuel, Andi Liu, Boyan Leng, Xiangning Chen and Zhongming Zhao
Epigenomes 2024, 8(2), 14; https://doi.org/10.3390/epigenomes8020014 - 01 Apr 2024
Abstract
►▼
Show Figures
Alzheimer’s Disease (AD) is a complex disease and the leading cause of dementia in older people. We aimed to uncover aspects of AD’s pathogenesis that may contribute to drug repurposing efforts by integrating DNA methylation and genetic data. Implementing the network-based tool, a
[...] Read more.
Alzheimer’s Disease (AD) is a complex disease and the leading cause of dementia in older people. We aimed to uncover aspects of AD’s pathogenesis that may contribute to drug repurposing efforts by integrating DNA methylation and genetic data. Implementing the network-based tool, a dense module search of genome-wide association studies (dmGWAS), we integrated a large-scale GWAS dataset with DNA methylation data to identify gene network modules associated with AD. Our analysis yielded 286 significant gene network modules. Notably, the foremost module included the BIN1 gene, showing the largest GWAS signal, and the GNAS gene, the most significantly hypermethylated. We conducted Web-based Cell-type-Specific Enrichment Analysis (WebCSEA) on genes within the top 10% of dmGWAS modules, highlighting monocyte as the most significant cell type (p < 5 × 10−12). Functional enrichment analysis revealed Gene Ontology Biological Process terms relevant to AD pathology (adjusted p < 0.05). Additionally, drug target enrichment identified five FDA-approved targets (p-value = 0.03) for further research. In summary, dmGWAS integration of genetic and epigenetic signals unveiled new gene interactions related to AD, offering promising avenues for future studies.
Full article
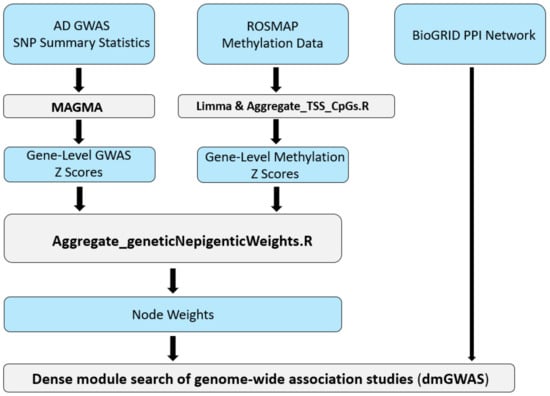
Figure 1
Open AccessReview
Pathogenesis of PM2.5-Related Disorders in Different Age Groups: Children, Adults, and the Elderly
by
Teerachai Amnuaylojaroen and Nichapa Parasin
Epigenomes 2024, 8(2), 13; https://doi.org/10.3390/epigenomes8020013 - 31 Mar 2024
Abstract
The effects of PM2.5 on human health fluctuate greatly among various age groups, influenced by a range of physiological and immunological reactions. This paper compares the pathogenesis of the disease caused by PM2.5 in people of different ages, focusing on how
[...] Read more.
The effects of PM2.5 on human health fluctuate greatly among various age groups, influenced by a range of physiological and immunological reactions. This paper compares the pathogenesis of the disease caused by PM2.5 in people of different ages, focusing on how children, adults, and the elderly are each susceptible to it because of differences in their bodies. Regarding children, exposure to PM2.5 is linked to many negative consequences. These factors consist of inflammation, oxidative stress, and respiratory problems, which might worsen pre-existing conditions and potentially cause neurotoxicity and developmental issues. Epigenetic changes can affect the immune system and make people more likely to get respiratory diseases. On the other hand, exposures during pregnancy can change how the cardiovascular and central nervous systems develop. In adults, the inhalation of PM2.5 is associated with a wide range of health problems. These include respiratory difficulties, reduced pulmonary function, and an increased susceptibility to illnesses such as asthma, chronic obstructive pulmonary disease (COPD), and lung cancer. In addition, exposure to PM2.5 induces systemic inflammation, cardiovascular diseases, insulin resistance, and neurotoxic consequences. Evident disturbances in the immune system and cognitive function demonstrate the broad impact of PM2.5. The elderly population is prone to developing respiratory and cardiovascular difficulties, which worsen their pre-existing health issues and raise the risk of cognitive decline and neurological illnesses. Having additional medical conditions, such as peptic ulcer disease, significantly increases the likelihood of being admitted to hospital.
Full article
(This article belongs to the Special Issue Environmental Epigenomes)
►▼
Show Figures
Figure 1
Open AccessArticle
Epigenetic Features in Newborns Associated with Preadolescence Lung Function and Asthma Acquisition during Adolescence
by
Mohammad Nahian Ferdous Abrar, Yu Jiang, Hongmei Zhang, Liang Li and Hasan Arshad
Epigenomes 2024, 8(2), 12; https://doi.org/10.3390/epigenomes8020012 - 22 Mar 2024
Abstract
The association between newborn DNA methylation (DNAm) and asthma acquisition (AA) during adolescence has been suggested. Lung function (LF) has been shown to be associated with asthma risk and its severity. However, the role of LF in the associations between DNAm and AA
[...] Read more.
The association between newborn DNA methylation (DNAm) and asthma acquisition (AA) during adolescence has been suggested. Lung function (LF) has been shown to be associated with asthma risk and its severity. However, the role of LF in the associations between DNAm and AA is unclear, and it is also unknown whether the association between DNAm and AA is consistent with that between DNAm and LF. We address this question through assessing newborn epigenetic features of preadolescence LF and of AA during adolescence, along with their biological pathways and processes. Our study’s primary medical significance lies in advancing the understanding of asthma’s early life origins. By investigating epigenetic markers in newborns and their association with lung function in preadolescence, we aim to uncover potential early biomarkers of asthma risk. This could facilitate earlier detection and intervention strategies. Additionally, exploring biological pathways linking early lung function to later asthma development can offer insights into the disease’s pathogenesis, potentially leading to novel therapeutic targets. Methods: The study was based on the Isle of Wight Birth cohort (IOWBC). Female subjects with DNAm data at birth and with no asthma at age 10 years were included (n = 249). The R package ttScreening was applied to identify CpGs potentially associated with AA from 10 to 18 years and with LF at age 10 (FEV1, FVC, and FEV1/FVC), respectively. Agreement in identified CpGs between AA and LF was examined, along with their biological pathways and processes via the R function gometh. We tested the findings in an independent cohort, the Avon Longitudinal Study of Parents and Children (ALSPAC), to examine overall replicability. Results: In IOWBC, 292 CpGs were detected with DNAm associated with AA and 1517 unique CpGs for LF (514 for FEV1, 436 for FVC, 408 for FEV1/FVC), with one overlapping CpG, cg23642632 (NCKAP1) between AA and LF. Among the IOWBC-identified CpGs, we further tested in ALSPAC and observed the highest agreement between the two cohorts in FVC with respect to the direction of association and statistical significance. Epigenetic enrichment analyses indicated non-specific connections in the biological pathways and processes between AA and LF. Conclusions: The present study suggests that FEV1, FVC, and FEV1/FVC (as objective measures of LF) and AA (incidence of asthma) are likely to have their own specific epigenetic features and biological pathways at birth. More replications are desirable to fully understand the complexity between DNAm, lung function, and asthma acquisition.
Full article
(This article belongs to the Special Issue Environmental Epigenomes)
►▼
Show Figures

Figure 1
Open AccessArticle
Opposite and Differently Altered Postmortem Changes in H3 and H3K9me3 Patterns in the Rat Frontal Cortex and Hippocampus
by
Karolina Dulka, Noémi Lajkó, Kálmán Nacsa and Karoly Gulya
Epigenomes 2024, 8(1), 11; https://doi.org/10.3390/epigenomes8010011 - 18 Mar 2024
Abstract
Temporal and spatial epigenetic modifications in the brain occur during ontogenetic development, pathophysiological disorders, and aging. When epigenetic marks, such as histone methylations, in brain autopsies or biopsy samples are studied, it is critical to understand their postmortem/surgical stability. For this study, the
[...] Read more.
Temporal and spatial epigenetic modifications in the brain occur during ontogenetic development, pathophysiological disorders, and aging. When epigenetic marks, such as histone methylations, in brain autopsies or biopsy samples are studied, it is critical to understand their postmortem/surgical stability. For this study, the frontal cortex and hippocampus of adult rats were removed immediately (controls) or after a postmortem delay of 15, 30, 60, 90, 120, or 150 min. The patterns of unmodified H3 and its trimethylated form H3K9me3 were analyzed in frozen samples for Western blot analysis and in formalin-fixed tissues embedded in paraffin for confocal microscopy. We found that both the unmodified H3 and H3K9me3 showed time-dependent but opposite changes and were altered differently in the frontal cortex and hippocampus with respect to postmortem delay. In the frontal cortex, the H3K9me3 marks increased approximately 450% with a slow parallel 20% decrease in the unmodified H3 histones after 150 min. In the hippocampus, the change was opposite, since H3K9me3 marks decreased steadily by approximately 65% after 150 min with a concomitant rapid increase of 20–25% in H3 histones at the same time. Confocal microscopy located H3K9me3 marks in the heterochromatic regions of the nuclei of all major cell types in the control brains: oligodendrocytes, astrocytes, neurons, and microglia. Therefore, epigenetic marks could be affected differently by postmortem delay in different parts of the brain.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures
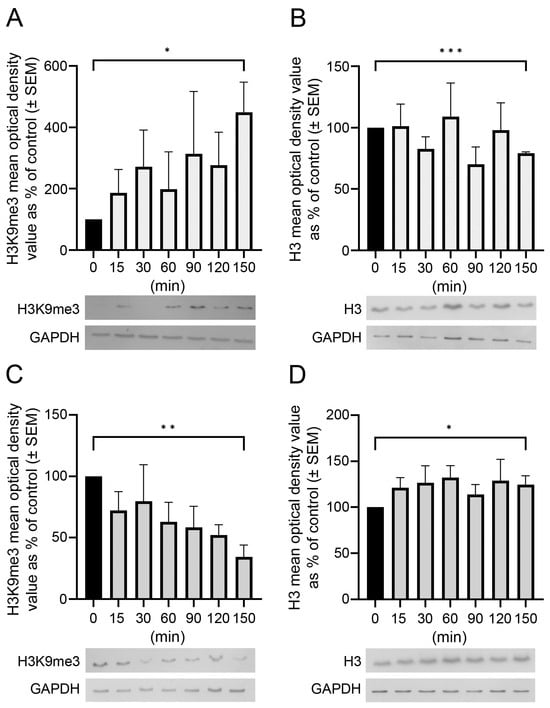
Figure 1
Open AccessArticle
Exploring Transcriptional Regulation of Beta Cell SASP by Brd4-Associated Proteins and Cell Cycle Control Protein p21
by
Jasmine Manji, Jasmine Pipella, Gabriel Brawerman and Peter J. Thompson
Epigenomes 2024, 8(1), 10; https://doi.org/10.3390/epigenomes8010010 - 06 Mar 2024
Abstract
Type 1 diabetes (T1D) is a metabolic disease resulting from progressive autoimmune destruction of insulin-producing pancreatic beta cells. Although the majority of beta cells are lost in T1D, a small subset undergoes senescence, a stress response involving growth arrest, DNA damage response, and
[...] Read more.
Type 1 diabetes (T1D) is a metabolic disease resulting from progressive autoimmune destruction of insulin-producing pancreatic beta cells. Although the majority of beta cells are lost in T1D, a small subset undergoes senescence, a stress response involving growth arrest, DNA damage response, and activation of a senescence-associated secretory phenotype (SASP). SASP in beta cells of the nonobese diabetic (NOD) mouse model of T1D and primary human islets is regulated at the level of transcription by bromodomain extra-terminal (BET) proteins, but the mechanisms remain unclear. To explore how SASP is transcriptionally regulated in beta cells, we used the NOD beta cell line NIT-1 to model beta cell SASP and identified binding partners of BET protein Brd4 and explored the role of the cyclin-dependent kinase inhibitor p21. Brd4 interacted with a variety of proteins in senescent NIT-1 cells including subunits of the Ino80 chromatin remodeling complex, which was expressed in beta cells during T1D progression in NOD mice and in human beta cells of control, autoantibody-positive, and T1D donors as determined from single-cell RNA-seq data. RNAi knockdown of p21 during senescence in NIT-1 cells did not significantly impact viability or SASP. Taken together, these results suggest that Brd4 interacts with several protein partners during senescence in NIT-1 cells, some of which may play roles in SASP gene activation and that p21 is dispensable for the SASP in this beta cell model.
Full article
(This article belongs to the Collection Epigenetic Mechanisms in Diabetes Research)
►▼
Show Figures

Figure 1
Open AccessReview
Dynamics and Epigenetics of the Epidermal Differentiation Complex
by
Wiesława Leśniak
Epigenomes 2024, 8(1), 9; https://doi.org/10.3390/epigenomes8010009 - 29 Feb 2024
Abstract
►▼
Show Figures
Epidermis is the outer skin layer built of specialized cells called keratinocytes. Keratinocytes undergo a unique differentiation process, also known as cornification, during which their gene expression pattern, morphology and other properties change remarkably to the effect that the terminally differentiated, cornified cells
[...] Read more.
Epidermis is the outer skin layer built of specialized cells called keratinocytes. Keratinocytes undergo a unique differentiation process, also known as cornification, during which their gene expression pattern, morphology and other properties change remarkably to the effect that the terminally differentiated, cornified cells can form a physical barrier, which separates the underlying tissues from the environment. Many genes encoding proteins that are important for epidermal barrier formation are located in a gene cluster called epidermal differentiation complex (EDC). Recent data provided valuable information on the dynamics of the EDC locus and the network of interactions between EDC gene promoters, enhancers and other regions, during keratinocytes differentiation. These data, together with results concerning changes in epigenetic modifications, provide a valuable insight into the mode of regulation of EDC gene expression.
Full article
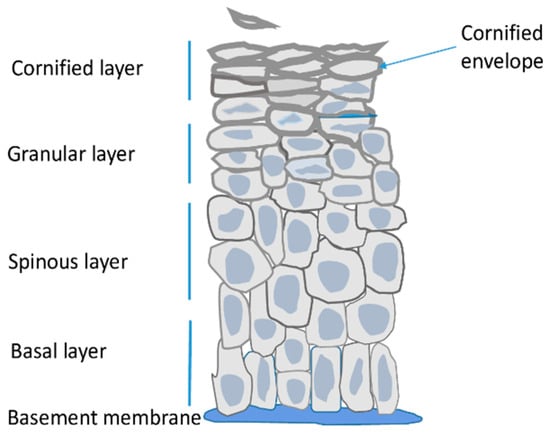
Figure 1
Open AccessReview
A Comparative Analysis of Mouse Imprinted and Random X-Chromosome Inactivation
by
Rebecca M. Malcore and Sundeep Kalantry
Epigenomes 2024, 8(1), 8; https://doi.org/10.3390/epigenomes8010008 - 10 Feb 2024
Abstract
The mammalian sexes are distinguished by the X and Y chromosomes. Whereas males harbor one X and one Y chromosome, females harbor two X chromosomes. To equalize X-linked gene expression between the sexes, therian mammals have evolved X-chromosome inactivation as a dosage compensation
[...] Read more.
The mammalian sexes are distinguished by the X and Y chromosomes. Whereas males harbor one X and one Y chromosome, females harbor two X chromosomes. To equalize X-linked gene expression between the sexes, therian mammals have evolved X-chromosome inactivation as a dosage compensation mechanism. During X-inactivation, most genes on one of the two X chromosomes in females are transcriptionally silenced, thus equalizing X-linked gene expression between the sexes. Two forms of X-inactivation characterize eutherian mammals, imprinted and random. Imprinted X-inactivation is defined by the exclusive inactivation of the paternal X chromosome in all cells, whereas random X-inactivation results in the silencing of genes on either the paternal or maternal X chromosome in individual cells. Both forms of X-inactivation have been studied intensively in the mouse model system, which undergoes both imprinted and random X-inactivation early in embryonic development. Stable imprinted and random X-inactivation requires the induction of the Xist long non-coding RNA. Following its induction, Xist RNA recruits proteins and complexes that silence genes on the inactive-X. In this review, we present a current understanding of the mechanisms of Xist RNA induction, and, separately, the establishment and maintenance of gene silencing on the inactive-X by Xist RNA during imprinted and random X-inactivation.
Full article
(This article belongs to the Special Issue X-Chromosome Inactivation)
►▼
Show Figures
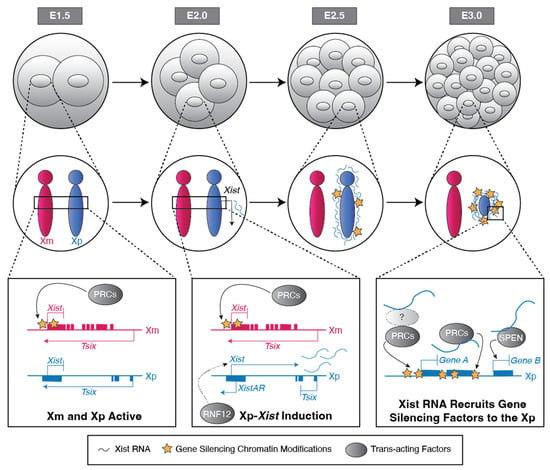
Figure 1
Open AccessReview
Targeting SWI/SNF Complexes in Cancer: Pharmacological Approaches and Implications
by
Megan R. Dreier, Jasmine Walia and Ivana L. de la Serna
Epigenomes 2024, 8(1), 7; https://doi.org/10.3390/epigenomes8010007 - 04 Feb 2024
Abstract
►▼
Show Figures
SWI/SNF enzymes are heterogeneous multi-subunit complexes that utilize the energy from ATP hydrolysis to remodel chromatin structure, facilitating transcription, DNA replication, and repair. In mammalian cells, distinct sub-complexes, including cBAF, ncBAF, and PBAF exhibit varying subunit compositions and have different genomic functions. Alterations
[...] Read more.
SWI/SNF enzymes are heterogeneous multi-subunit complexes that utilize the energy from ATP hydrolysis to remodel chromatin structure, facilitating transcription, DNA replication, and repair. In mammalian cells, distinct sub-complexes, including cBAF, ncBAF, and PBAF exhibit varying subunit compositions and have different genomic functions. Alterations in the SWI/SNF complex and sub-complex functions are a prominent feature in cancer, making them attractive targets for therapeutic intervention. Current strategies in cancer therapeutics involve the use of pharmacological agents designed to bind and disrupt the activity of SWI/SNF complexes or specific sub-complexes. Inhibitors targeting the catalytic subunits, SMARCA4/2, and small molecules binding SWI/SNF bromodomains are the primary approaches for suppressing SWI/SNF function. Proteolysis-targeting chimeras (PROTACs) were generated by the covalent linkage of the bromodomain or ATPase-binding ligand to an E3 ligase-binding moiety. This engineered connection promotes the degradation of specific SWI/SNF subunits, enhancing and extending the impact of this pharmacological intervention in some cases. Extensive preclinical studies have underscored the therapeutic potential of these drugs across diverse cancer types. Encouragingly, some of these agents have progressed from preclinical research to clinical trials, indicating a promising stride toward the development of effective cancer therapeutics targeting SWI/SNF complex and sub-complex functions.
Full article

Figure 1
Open AccessReview
Orchestrating Asymmetric Expression: Mechanisms behind Xist Regulation
by
Samuel Jesus Luchsinger-Morcelle, Joost Gribnau and Hegias Mira-Bontenbal
Epigenomes 2024, 8(1), 6; https://doi.org/10.3390/epigenomes8010006 - 01 Feb 2024
Abstract
Compensation for the gene dosage disequilibrium between sex chromosomes in mammals is achieved in female cells by repressing one of its X chromosomes through a process called X chromosome inactivation (XCI), exemplifying the control of gene expression by epigenetic mechanisms. A critical player
[...] Read more.
Compensation for the gene dosage disequilibrium between sex chromosomes in mammals is achieved in female cells by repressing one of its X chromosomes through a process called X chromosome inactivation (XCI), exemplifying the control of gene expression by epigenetic mechanisms. A critical player in this mechanism is Xist, a long, non-coding RNA upregulated from a single X chromosome during early embryonic development in female cells. Over the past few decades, many factors involved at different levels in the regulation of Xist have been discovered. In this review, we hierarchically describe and analyze the different layers of Xist regulation operating concurrently and intricately interacting with each other to achieve asymmetric and monoallelic upregulation of Xist in murine female cells. We categorize these into five different classes: DNA elements, transcription factors, other regulatory proteins, long non-coding RNAs, and the chromatin and topological landscape surrounding Xist.
Full article
(This article belongs to the Special Issue X-Chromosome Inactivation)
►▼
Show Figures

Figure 1
Open AccessArticle
Angio-Long Noncoding RNA MALAT1 (rs3200401) and MIAT (rs1061540) Gene Variants in Ovarian Cancer
by
Manal S. Fawzy, Afaf T. Ibrahiem, Dalia Mohammad Osman, Amany I. Almars, Maali Subhi Alshammari, Layan Tariq Almazyad, Noof Daif Allah Almatrafi, Renad Tariq Almazyad and Eman A. Toraih
Epigenomes 2024, 8(1), 5; https://doi.org/10.3390/epigenomes8010005 - 29 Jan 2024
Abstract
►▼
Show Figures
The genotyping of long non-coding RNA (lncRNA)-related single-nucleotide polymorphisms (SNPs) could be associated with cancer risk and/or progression. This study aimed to analyze the angiogenesis-related lncRNAs MALAT1 (rs3200401) and MIAT (rs1061540) variants in patients with ovarian cancer (OC) using “Real-Time allelic discrimination polymerase
[...] Read more.
The genotyping of long non-coding RNA (lncRNA)-related single-nucleotide polymorphisms (SNPs) could be associated with cancer risk and/or progression. This study aimed to analyze the angiogenesis-related lncRNAs MALAT1 (rs3200401) and MIAT (rs1061540) variants in patients with ovarian cancer (OC) using “Real-Time allelic discrimination polymerase chain reaction” in 182 formalin-fixed paraffin-embedded (FFPE) samples of benign, borderline, and primary malignant ovarian tissues. Differences in the genotype frequencies between low-grade ovarian epithelial tumors (benign/borderline) and malignant tumors and between high-grade malignant epithelial tumors and malignant epithelial tumors other than high-grade serous carcinomas were compared. Odds ratios (ORs)/95% confidence intervals were calculated as measures of the association strength. Additionally, associations of the genotypes with the available pathological data were analyzed. The heterozygosity of MALAT1 rs3200401 was the most common genotype (47.8%), followed by C/C (36.3%). Comparing the study groups, no significant differences were observed regarding this variant. In contrast, the malignant epithelial tumors had a higher frequency of the MIAT rs1061540 C/C genotype compared to the low-grade epithelial tumor cohorts (56.7% vs. 37.6, p = 0.031). The same genotype was significantly higher in high-grade serous carcinoma than its counterparts (69.4% vs. 43.8%, p = 0.038). Multivariate Cox regression analysis showed that the age at diagnosis was significantly associated with the risk of OC development. In contrast, the MIAT T/T genotype was associated with a low risk of malignant epithelial tumors under the homozygote comparison model (OR = 0.37 (0.16–0.83), p = 0.017). Also, MIAT T allele carriers were less likely to develop high-grade serous carcinoma under heterozygote (CT vs. CC; OR = 0.33 (0.12–0.88), p = 0.027) and homozygote (TT vs. CC; OR = 0.26 (0.07–0.90), p = 0.034) comparison models. In conclusion, our data provide novel evidence for a potential association between the lncRNA MIAT rs1061540 and the malignant condition of ovarian cancer, suggesting the involvement of such lncRNAs in OC development.
Full article
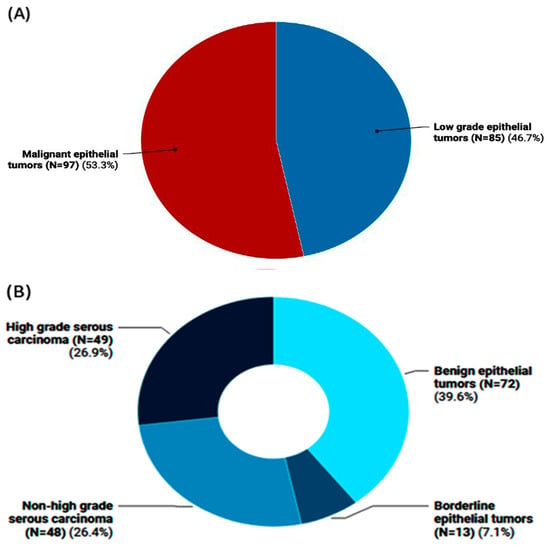
Figure 1
Open AccessArticle
Epigenetics of Genes Preferentially Expressed in Dissimilar Cell Populations: Myoblasts and Cerebellum
by
Melanie Ehrlich, Kenneth C. Ehrlich, Michelle Lacey, Carl Baribault, Sagnik Sen, Pierre-Olivier Estève and Sriharsa Pradhan
Epigenomes 2024, 8(1), 4; https://doi.org/10.3390/epigenomes8010004 - 26 Jan 2024
Abstract
While studying myoblast methylomes and transcriptomes, we found that CDH15 had a remarkable preference for expression in both myoblasts and cerebellum. To understand how widespread such a relationship was and its epigenetic and biological correlates, we systematically looked for genes with similar transcription
[...] Read more.
While studying myoblast methylomes and transcriptomes, we found that CDH15 had a remarkable preference for expression in both myoblasts and cerebellum. To understand how widespread such a relationship was and its epigenetic and biological correlates, we systematically looked for genes with similar transcription profiles and analyzed their DNA methylation and chromatin state and accessibility profiles in many different cell populations. Twenty genes were expressed preferentially in myoblasts and cerebellum (Myob/Cbl genes). Some shared DNA hypo- or hypermethylated regions in myoblasts and cerebellum. Particularly striking was ZNF556, whose promoter is hypomethylated in expressing cells but highly methylated in the many cell populations that do not express the gene. In reporter gene assays, we demonstrated that its promoter’s activity is methylation sensitive. The atypical epigenetics of ZNF556 may have originated from its promoter’s hypomethylation and selective activation in sperm progenitors and oocytes. Five of the Myob/Cbl genes (KCNJ12, ST8SIA5, ZIC1, VAX2, and EN2) have much higher RNA levels in cerebellum than in myoblasts and displayed myoblast-specific hypermethylation upstream and/or downstream of their promoters that may downmodulate expression. Differential DNA methylation was associated with alternative promoter usage for Myob/Cbl genes MCF2L, DOK7, CNPY1, and ANK1. Myob/Cbl genes PAX3, LBX1, ZNF556, ZIC1, EN2, and VAX2 encode sequence-specific transcription factors, which likely help drive the myoblast and cerebellum specificity of other Myob/Cbl genes. This study extends our understanding of epigenetic/transcription associations related to differentiation and may help elucidate relationships between epigenetic signatures and muscular dystrophies or cerebellar-linked neuropathologies.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures
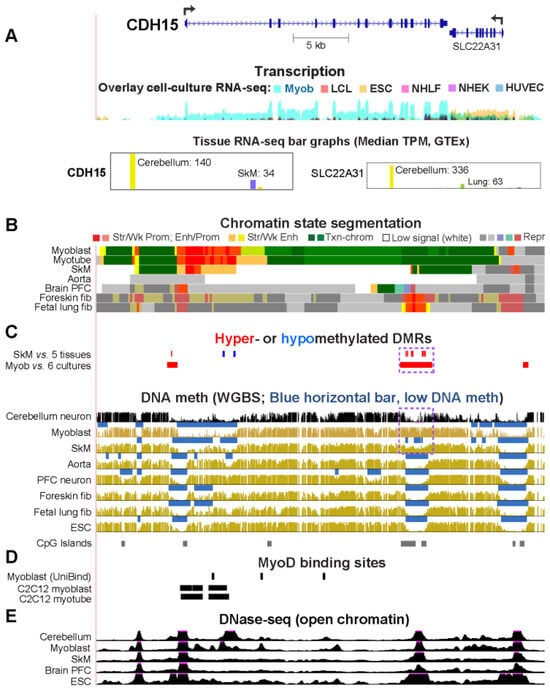
Figure 1
Open AccessArticle
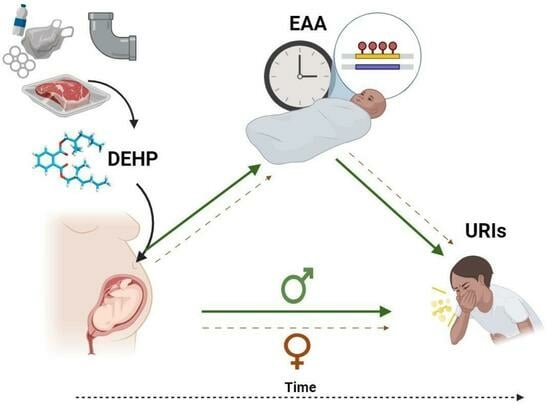
Sex-Specific Associations between Prenatal Exposure to Di(2-ethylhexyl) Phthalate, Epigenetic Age Acceleration, and Susceptibility to Early Childhood Upper Respiratory Infections
by
Sarah M. Merrill, Nicole Letourneau, Gerald F. Giesbrecht, Karlie Edwards, Julia L. MacIsaac, Jonathan W. Martin, Amy M. MacDonald, David W. Kinniburgh, Michael S. Kobor, Deborah Dewey, Gillian England-Mason and The APrON Study Team
Epigenomes 2024, 8(1), 3; https://doi.org/10.3390/epigenomes8010003 - 26 Jan 2024
Abstract
Di(2-ethylhexyl) phthalate (DEHP) is a common plasticizer that can affect immune system development and susceptibility to infection. Aging processes (measured as epigenetic age acceleration (EAA)) may mediate the immune-related effects of prenatal exposure to DEHP. This study’s objective was to examine associations between
[...] Read more.
Di(2-ethylhexyl) phthalate (DEHP) is a common plasticizer that can affect immune system development and susceptibility to infection. Aging processes (measured as epigenetic age acceleration (EAA)) may mediate the immune-related effects of prenatal exposure to DEHP. This study’s objective was to examine associations between prenatal DEHP exposure, EAA at three months of age, and the number of upper respiratory infections (URIs) from 12 to 18 months of age using a sample of 69 maternal–child pairs from a Canadian pregnancy cohort. Blood DNA methylation data were generated using the Infinium HumanMethylation450 BeadChip; EAA was estimated using Horvath’s pan-tissue clock. Robust regressions examined overall and sex-specific associations. Higher prenatal DEHP exposure (B = 6.52, 95% CI = 1.22, 11.81) and increased EAA (B = 2.98, 95% CI = 1.64, 4.32) independently predicted more URIs. In sex-specific analyses, some similar effects were noted for boys, and EAA mediated the association between prenatal DEHP exposure and URIs. In girls, higher prenatal DEHP exposure was associated with decreased EAA, and no mediation was noted. Higher prenatal DEHP exposure may be associated with increased susceptibility to early childhood URIs, particularly in boys, and aging biomarkers such as EAA may be a biological mechanism. Larger cohort studies examining the potential developmental immunotoxicity of phthalates are needed.
Full article
(This article belongs to the Collection Feature Papers in Epigenomes)
►▼
Show Figures
Graphical abstract
Open AccessReview
The Genomic Shock Hypothesis: Genetic and Epigenetic Alterations of Transposable Elements after Interspecific Hybridization in Plants
by
Carlos de Tomás and Carlos M. Vicient
Epigenomes 2024, 8(1), 2; https://doi.org/10.3390/epigenomes8010002 - 27 Dec 2023
Abstract
Transposable elements (TEs) are major components of plant genomes with the ability to change their position in the genome or to create new copies of themselves in other positions in the genome. These can cause gene disruption and large-scale genomic alterations, including inversions,
[...] Read more.
Transposable elements (TEs) are major components of plant genomes with the ability to change their position in the genome or to create new copies of themselves in other positions in the genome. These can cause gene disruption and large-scale genomic alterations, including inversions, deletions, and duplications. Host organisms have evolved a set of mechanisms to suppress TE activity and counter the threat that they pose to genome integrity. These includes the epigenetic silencing of TEs mediated by a process of RNA-directed DNA methylation (RdDM). In most cases, the silencing machinery is very efficient for the vast majority of TEs. However, there are specific circumstances in which TEs can evade such silencing mechanisms, for example, a variety of biotic and abiotic stresses or in vitro culture. Hybridization is also proposed as an inductor of TE proliferation. In fact, the discoverer of the transposons, Barbara McClintock, first hypothesized that interspecific hybridization provides a “genomic shock” that inhibits the TE control mechanisms leading to the mobilization of TEs. However, the studies carried out on this topic have yielded diverse results, showing in some cases a total absence of mobilization or being limited to only some TE families. Here, we review the current knowledge about the impact of interspecific hybridization on TEs in plants and the possible implications of changes in the epigenetic mechanisms.
Full article
(This article belongs to the Collection Epigenetic Control in Plants)
►▼
Show Figures
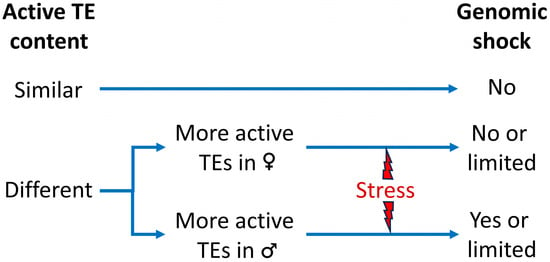
Figure 1
Open AccessReview
Inheritance of Stress Responses via Small Non-Coding RNAs in Invertebrates and Mammals
by
Maria C. Ow and Sarah E. Hall
Epigenomes 2024, 8(1), 1; https://doi.org/10.3390/epigenomes8010001 - 19 Dec 2023
Cited by 1
Abstract
While reports on the generational inheritance of a parental response to stress have been widely reported in animals, the molecular mechanisms behind this phenomenon have only recently emerged. The booming interest in epigenetic inheritance has been facilitated in part by the discovery that
[...] Read more.
While reports on the generational inheritance of a parental response to stress have been widely reported in animals, the molecular mechanisms behind this phenomenon have only recently emerged. The booming interest in epigenetic inheritance has been facilitated in part by the discovery that small non-coding RNAs are one of its principal conduits. Discovered 30 years ago in the Caenorhabditis elegans nematode, these small molecules have since cemented their critical roles in regulating virtually all aspects of eukaryotic development. Here, we provide an overview on the current understanding of epigenetic inheritance in animals, including mice and C. elegans, as it pertains to stresses such as temperature, nutritional, and pathogenic encounters. We focus on C. elegans to address the mechanistic complexity of how small RNAs target their cohort mRNAs to effect gene expression and how they govern the propagation or termination of generational perdurance in epigenetic inheritance. Presently, while a great amount has been learned regarding the heritability of gene expression states, many more questions remain unanswered and warrant further investigation.
Full article
(This article belongs to the Special Issue Transgenerational Epigenetic Inheritance)
►▼
Show Figures
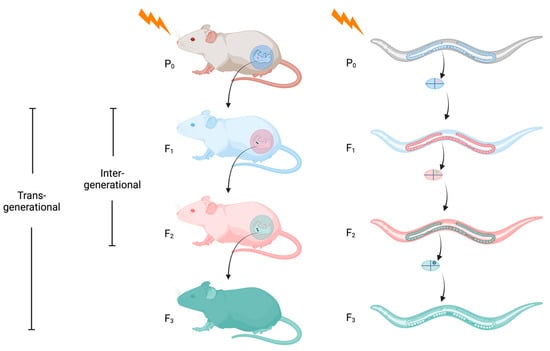
Figure 1
Open AccessReview
Epigenetic Mechanisms in Hematologic Aging and Premalignant Conditions
by
Bowen Yan, Qingchen Yuan and Olga A. Guryanova
Epigenomes 2023, 7(4), 32; https://doi.org/10.3390/epigenomes7040032 - 12 Dec 2023
Cited by 1
Abstract
Hematopoietic stem cells (HSCs) are essential for maintaining overall health by continuously generating blood cells throughout an individual’s lifespan. However, as individuals age, the hematopoietic system undergoes significant functional decline, rendering them more susceptible to age-related diseases. Growing research evidence has highlighted the
[...] Read more.
Hematopoietic stem cells (HSCs) are essential for maintaining overall health by continuously generating blood cells throughout an individual’s lifespan. However, as individuals age, the hematopoietic system undergoes significant functional decline, rendering them more susceptible to age-related diseases. Growing research evidence has highlighted the critical role of epigenetic regulation in this age-associated decline. This review aims to provide an overview of the diverse epigenetic mechanisms involved in the regulation of normal HSCs during the aging process and their implications in aging-related diseases. Understanding the intricate interplay of epigenetic mechanisms that contribute to aging-related changes in the hematopoietic system holds great potential for the development of innovative strategies to delay the aging process. In fact, interventions targeting epigenetic modifications have shown promising outcomes in alleviating aging-related phenotypes and extending lifespan in various animal models. Small molecule-based therapies and reprogramming strategies enabling epigenetic rejuvenation have emerged as effective approaches for ameliorating or even reversing aging-related conditions. By acquiring a deeper understanding of these epigenetic mechanisms, it is anticipated that interventions can be devised to prevent or mitigate the rates of hematologic aging and associated diseases later in life. Ultimately, these advancements have the potential to improve overall health and enhance the quality of life in aging individuals.
Full article
(This article belongs to the Special Issue Epigenetics in Hematologic Malignancies)
►▼
Show Figures
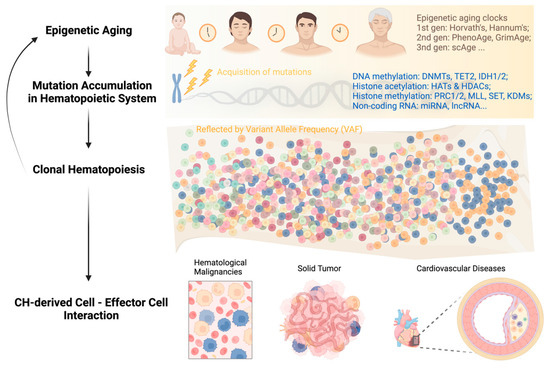
Figure 1
Open AccessReview
World Trade Center Exposure, DNA Methylation Changes, and Cancer: A Review of Current Evidence
by
Stephanie Tuminello, Emelie Nguyen, Nedim Durmus, Ramazan Alptekin, Muhammed Yilmaz, Maria Cecilia Crisanti, Matija Snuderl, Yu Chen, Yongzhao Shao, Joan Reibman, Emanuela Taioli and Alan A. Arslan
Epigenomes 2023, 7(4), 31; https://doi.org/10.3390/epigenomes7040031 - 08 Dec 2023
Abstract
Introduction: Known carcinogens in the dust and fumes from the destruction of the World Trade Center (WTC) towers on 9 November 2001 included metals, asbestos, and organic pollutants, which have been shown to modify epigenetic status. Epigenome-wide association analyses (EWAS) using uniform
[...] Read more.
Introduction: Known carcinogens in the dust and fumes from the destruction of the World Trade Center (WTC) towers on 9 November 2001 included metals, asbestos, and organic pollutants, which have been shown to modify epigenetic status. Epigenome-wide association analyses (EWAS) using uniform (Illumina) methodology have identified novel epigenetic profiles of WTC exposure. Methods: We reviewed all published data, comparing differentially methylated gene profiles identified in the prior EWAS studies of WTC exposure. This included DNA methylation changes in blood-derived DNA from cases of cancer-free “Survivors” and those with breast cancer, as well as tissue-derived DNA from “Responders” with prostate cancer. Emerging molecular pathways related to the observed DNA methylation changes in WTC-exposed groups were explored and summarized. Results: WTC dust exposure appears to be associated with DNA methylation changes across the genome. Notably, WTC dust exposure appears to be associated with increased global DNA methylation; direct dysregulation of cancer genes and pathways, including inflammation and immune system dysregulation; and endocrine system disruption, as well as disruption of cholesterol homeostasis and lipid metabolism. Conclusion: WTC dust exposure appears to be associated with biologically meaningful DNA methylation changes, with implications for carcinogenesis and development of other chronic diseases.
Full article
(This article belongs to the Special Issue Environmental Epigenomes)
Open AccessArticle
Comparison of Yersinia enterocolitica DNA Methylation at Ambient and Host Temperatures
by
Dustin J. Van Hofwegen, Carolyn J. Hovde and Scott A. Minnich
Epigenomes 2023, 7(4), 30; https://doi.org/10.3390/epigenomes7040030 - 30 Nov 2023
Abstract
Pathogenic bacteria recognize environmental cues to vary gene expression for host adaptation. Moving from ambient to host temperature, Yersinia enterocolitica responds by immediately repressing flagella synthesis and inducing the virulence plasmid (pYV)-encoded type III secretion system. In contrast, shifting from host to ambient temperature
[...] Read more.
Pathogenic bacteria recognize environmental cues to vary gene expression for host adaptation. Moving from ambient to host temperature, Yersinia enterocolitica responds by immediately repressing flagella synthesis and inducing the virulence plasmid (pYV)-encoded type III secretion system. In contrast, shifting from host to ambient temperature requires 2.5 generations to restore motility, suggesting a link to the cell cycle. We hypothesized that differential DNA methylation contributes to temperature-regulated gene expression. We tested this hypothesis by comparing single-molecule real-time (SMRT) sequencing of Y. enterocolitica DNA from cells growing exponentially at 22 °C and 37 °C. The inter-pulse duration ratio rather than the traditional QV scoring was the kinetic metric to compare DNA from cells grown at each temperature. All 565 YenI restriction sites were fully methylated at both temperatures. Among the 27,118 DNA adenine methylase (Dam) sites, 42 had differential methylation patterns, while 17 remained unmethylated regardless of the temperature. A subset of the differentially methylated Dam sites localized to promoter regions of predicted regulatory genes including LysR-type and PadR-like transcriptional regulators and a cyclic-di-GMP phosphodiesterase. The unmethylated Dam sites localized with a bias to the replication terminus, suggesting they were protected from Dam methylase. No cytosine methylation was detected at Dcm sites.
Full article
(This article belongs to the Special Issue Epigenetic and Epitranscriptomic Determinants of Host-Microbe Interactions)
►▼
Show Figures
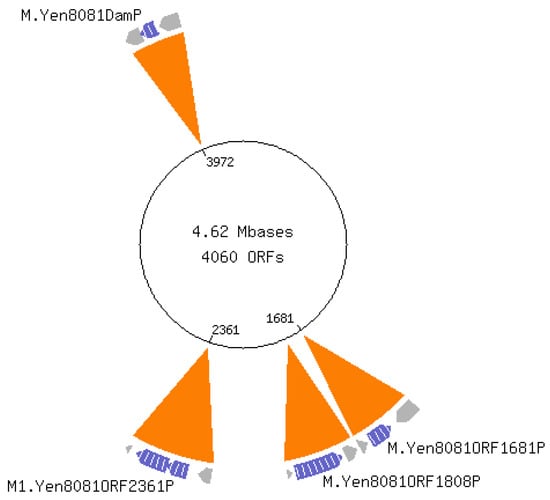
Figure 1
Open AccessReview
Out of the Silence: Insights into How Genes Escape X-Chromosome Inactivation
by
Samantha B. Peeters, Bronwyn J. Posynick and Carolyn J. Brown
Epigenomes 2023, 7(4), 29; https://doi.org/10.3390/epigenomes7040029 - 23 Nov 2023
Cited by 1
Abstract
The silencing of all but one X chromosome in mammalian cells is a remarkable epigenetic process leading to near dosage equivalence in X-linked gene products between the sexes. However, equally remarkable is the ability of a subset of genes to continue to be
[...] Read more.
The silencing of all but one X chromosome in mammalian cells is a remarkable epigenetic process leading to near dosage equivalence in X-linked gene products between the sexes. However, equally remarkable is the ability of a subset of genes to continue to be expressed from the otherwise inactive X chromosome—in some cases constitutively, while other genes are variable between individuals, tissues or cells. In this review we discuss the advantages and disadvantages of the approaches that have been used to identify escapees. The identity of escapees provides important clues to mechanisms underlying escape from XCI, an arena of study now moving from correlation to functional studies. As most escapees show greater expression in females, the not-so-inactive X chromosome is a substantial contributor to sex differences in humans, and we highlight some examples of such impact.
Full article
(This article belongs to the Special Issue X-Chromosome Inactivation)
►▼
Show Figures

Graphical abstract

Journal Menu
► ▼ Journal Menu-
- Epigenomes Home
- Aims & Scope
- Editorial Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Topical Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
Special Issues
Special Issue in
Epigenomes
RNA Modification in Inflammation and Metabolism
Guest Editors: Juncheng Wei, Cristina Espinosa-Diez, Kezhong ZhangDeadline: 30 April 2024
Special Issue in
Epigenomes
A Commemorative Issue in Honor of the 30th Anniversary of the Epigenetics Society
Guest Editors: Humaira Gowher, Assam El-Osta, Andrea FusoDeadline: 31 December 2024
Topical Collections
Topical Collection in
Epigenomes
Epigenetic Mechanisms in Diabetes Research
Collection Editor: Assam El-Osta
Topical Collection in
Epigenomes
Feature Papers in Epigenomes
Collection Editors: Che-Kun James Shen, Ivana De la Serna





