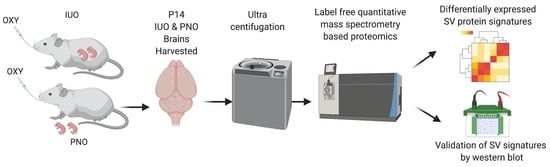
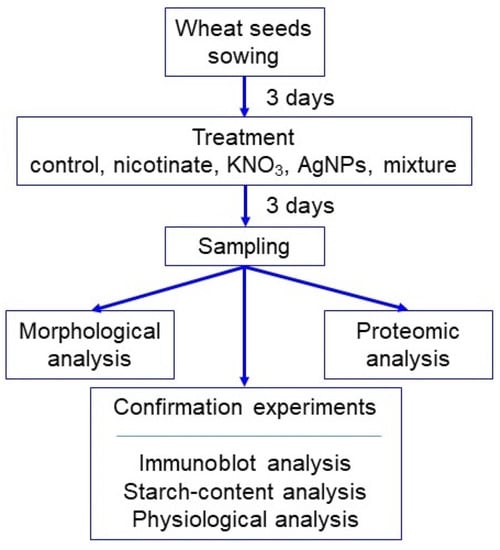
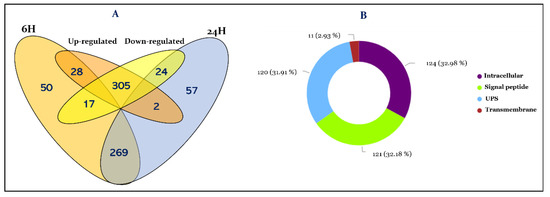
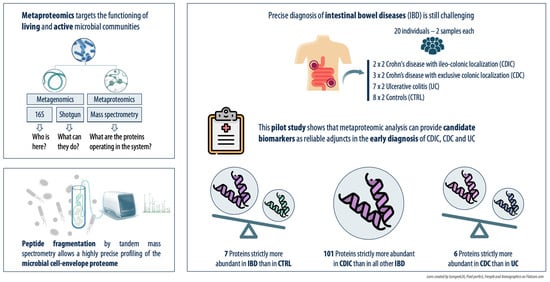
Deciphering the Proteome in Cell Biology and Diseases
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Cell Methods".
Viewed by 51097Editor
Interests: proteomics; glycoproteomics; PTM characterization; mass spectrometry; pancreatic cancer
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleague,
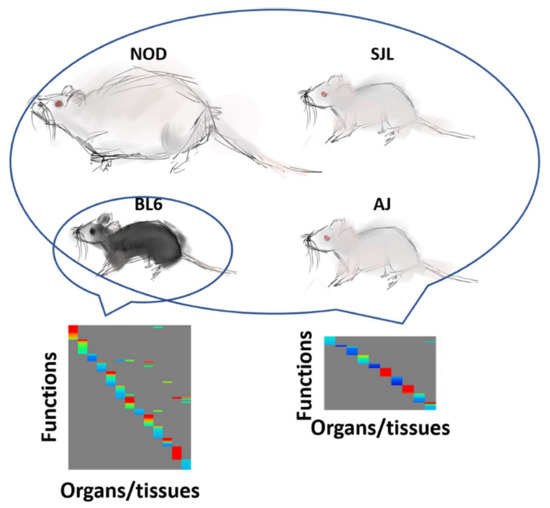
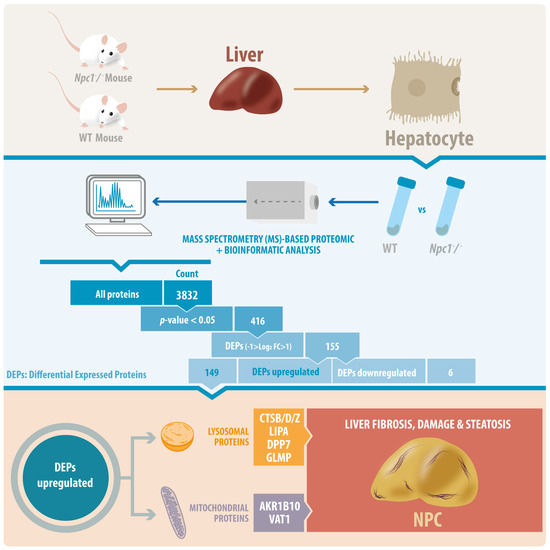
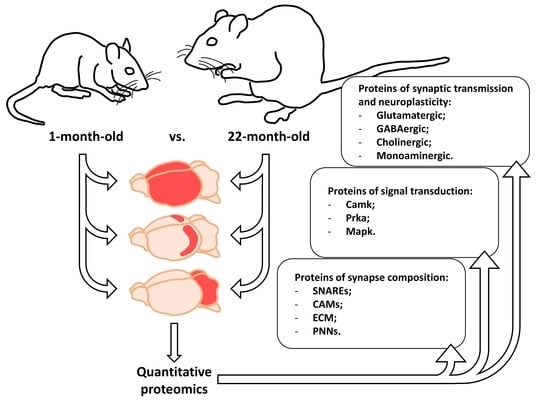
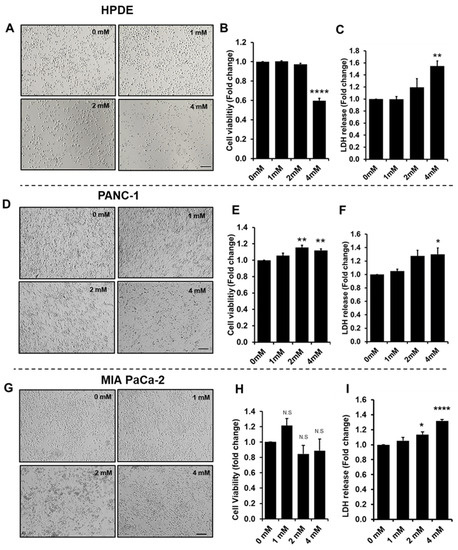
Proteins are essential functional biomolecules that participate in almost all cellular processes, and are profoundly implicated in disease settings. They form a highly organized and structured proteome that orchestrates synthetic, catalytic, and regulatory functions in the cell. The constituents of the cellular proteome, including protein identity, abundance, structure, post-translational modifications (PTMs), polymorphism, and cellular localization, dynamically and quantitatively define the phenotype and functional state of a cell. With the advances in mass spectrometry, bioinformatics, and knowledge base, proteomics has become a pivotal platform enabling the systematic study of the proteome and its functional implications in cell biology and diseases. While challenges still remain, the current state of proteomics makes it possible not only to conduct comprehensive global proteome profiling, but also hypothesis-driven targeted analysis and sophisticated site- and structure-specific PTM characterization, as well as meta-approaches for microbiome analysis. From the study of molecular mechanisms at cellular, organelle, and other subcellular levels to the characterization of various clinical specimens for facilitating disease treatment and biomarker discovery, proteomics—as part of systems biology endeavors—has been driving many new discoveries and hypotheses in the study of cell biology and disease. In this Topical Collection, we welcome the submission of original research or review articles aiming at the broad field of basic, translational, and clinical research using proteomic approaches.
Dr. Sheng Pan
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- proteomics
- mass spectrometry
- post-translational modifications
- bioinformatics
- biomarker
- proteome