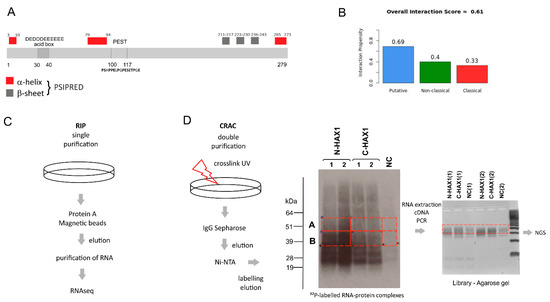
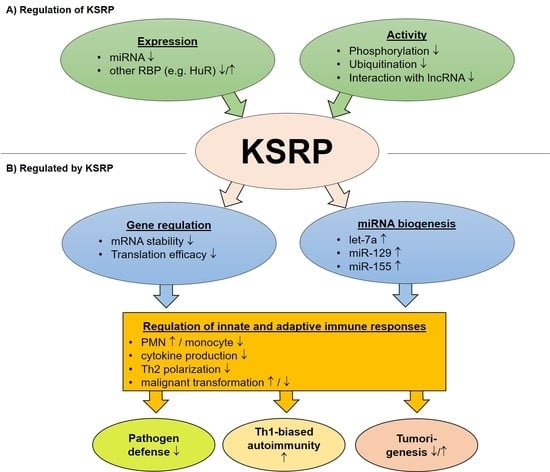
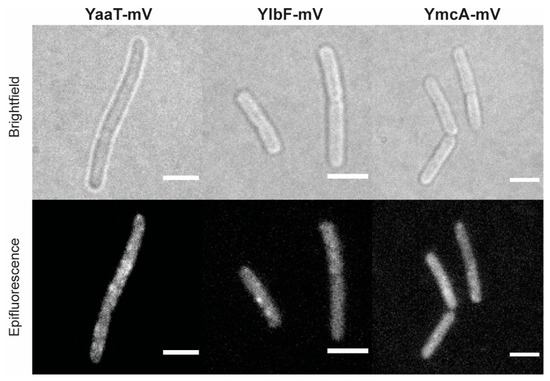
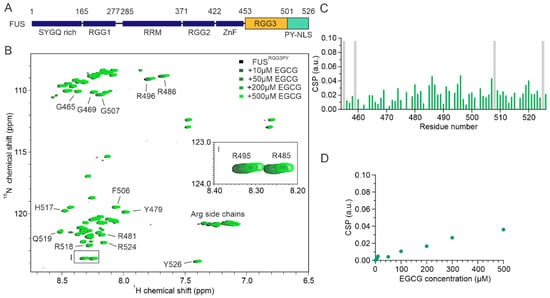
The Complex World of RNA-Protein Complexes—Emerging Players in Cellular Networks
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Cellular Biophysics".
Viewed by 28827Editor
Interests: protein-RNA interactions; histone methylation; liquid-liquid phase separation; intrinsically disordered proteins; histone lysine methyltransferases; long noncoding RNAs; RNA structure
Topical Collection Information
Dear Colleagues,
RNAs are involved in almost every cellular process—from gene expression regulation to molecular chaperoning. They are capable of forming various interactions with other RNA molecules, proteins, and DNA, which enables them to play central roles in the formation of cellular networks. Central to their proper function is the correct recognition of their native partners and the proper regulation of binding. Accumulating evidence shows that the RNA structure is essential in the molecular recognition of RNA binding proteins, and the immense effort invested in identifying the RNA–protein interaction networks of cells has produced an ever-growing amount of information on the structure–function relationship of RNAs and RNA–protein complexes. A clear understanding of the nature of RNA–protein binding is further complicated by the nonconventional interactions between RNA and protein that induce phase separation and the formation of membraneless organelles. Urgent questions regarding the recognition specificity, regulation of binding, and structural determinants of such processes from both RNA and protein must be elucidated.
Given the crucial roles of these interactions in the development of several diseases, RNAs are becoming attractive targets for drug development, offering new strategies to tackle difficult medical challenges; however, effective targeting requires detailed structural knowledge, which is relatively limited to date.
The aim of this Topical Collection is to provide an opportunity for researchers to present their latest results in the field of RNA–protein interactions in relation to the regulation of various cellular processes, and also to offer summaries on the most recent developments in the research field.
We look forward to your contribution.
Dr. Ágnes Tantos
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- RNA structure
- RNA-protein complex
- drug design
- RNA biomarkers
- membraneless organelles
- non-coding RNA
- RNA binding protein
- RNA interactions