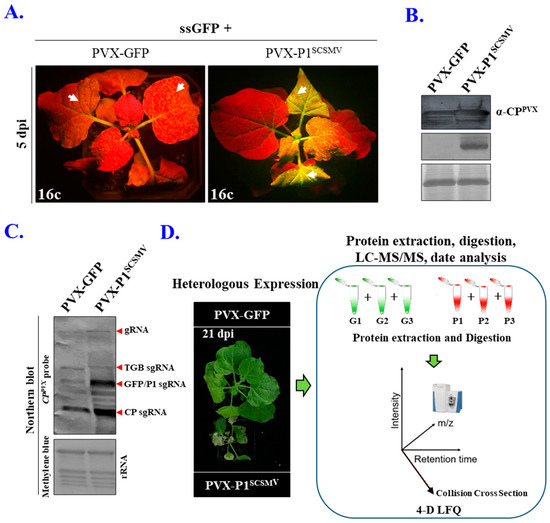
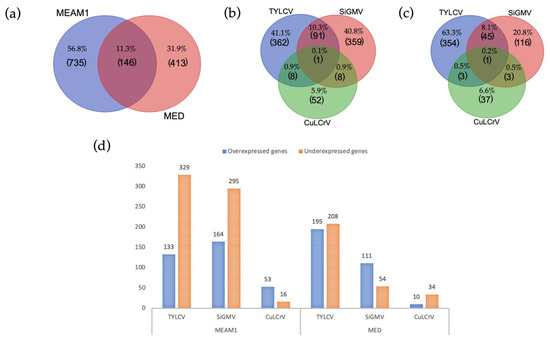
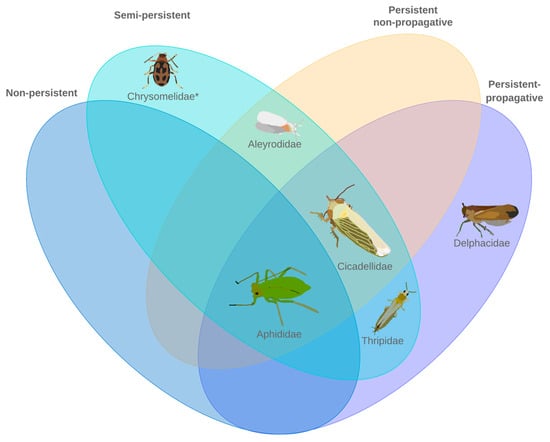
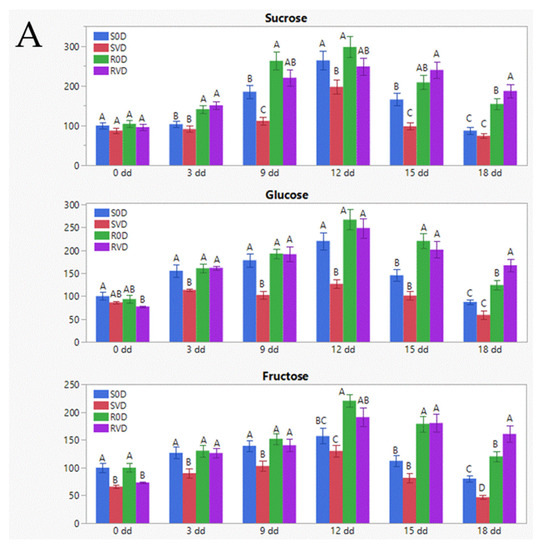
Plant-Virus/Viroid-Vector Interactions (Closed)
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Plant, Algae and Fungi Cell Biology".
Viewed by 49945Editors
Interests: viroids; viroid diseases; mycoviroids; plant viruses; molecular virology; molecular biology; genomics; origin; evolution
Special Issues, Collections and Topics in MDPI journals
Interests: plant–virus–insect interactions; plant stress responses; resistance to plant viruses
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Viruses were discovered as a new class of pathogens at the end of the 19th century. The iconic tobacco mosaic virus played a predominant role in that discovery. In the mid-1950s, it was discovered that the genetic information controlling viral replication in infected cells is carried in the nucleic acid core of each virus particle and that complete infectious particles are reconstituted from the protein and RNA. In 1973, one of the guest editors (A.H.) demonstrated that RNA viruses encode virus-specific RNA-dependent RNA polymerase (RdRp), known as “replicase”, to carry out their fully RNA-based genome replication. Later, others showed that RdRp is also involved in transcription. Until 1967, all known plant viruses were RNA viruses and the first DNA plant virus was discovered in 1968. Viroids were discovered in 1971. Viroids, the smallest known infectious agents, comprise a novel class of infectious RNA that replicates autonomously in infected cells, exists in circular and linear forms, and does not code for proteins. They belong to the new order of subviral agents. Some viruses and viroids may be transmitted by insects and other vectors. Plant viruses and viroids have had an impact on the science of virology, plant pathology, botany, microbiology, entomology, molecular biology, and breeding for resistance.
Advances in basic and applied research on Plant–Virus/Viroid–Vector Interactions have contributed to our understanding of and provided clear insight into the cellular localization, biology, genomics, genetics, molecular biology, molecular evolution, molecular ecology, biochemistry, and biophysics of the associated plant–pathogen interactions.
This Special Issue’s objective is to present a collection of original research and review articles describing recent advances and developments in Plant–Virus/Viroid–Vector Interactions. Topics of particular interest to this Special Issue include, but are not limited to:
-molecular analysis of factors or components that affect or modulate plant–virus/viroid–vector interactions;
-genetic, evolution, epidemiology, and ecology studies that contribute significantly to our understanding of plant–virus/viroid interactions and plant–virus/viroid–vector interactions
- the molecular evolution and molecular ecology of interactions between plants and viruses/viroids and eventual vectors; and
-advances in methods for studying the molecular or non-molecular aspects of plant–virus/viroid and plant–virus/viroid–vector interactions.
Dr. Ahmed Hadidi
Emeritus Lead Scientist
Guest Editor
Prof. Henryk Hanokh Czosnek
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- plant viruses
- RNA viruses
- DNA viruses
- virus diseases
- virus-resistant genes
- virus evolution
- viroids
- viroid diseases
- viroid evolution
- nucleus
- cytoplasm
- transcriptome
- pathogenesis-related (PR) proteins
- virus or viroid-induced gene silencing
- genome editing
- next-generation sequencing
- microarray profiling
- recombinant DNA technology
- transgenic plants
- insect vectors
- molecularly assisted breeding for resistance