Cancer Biomarkers in Body Fluids
A topical collection in Cancers (ISSN 2072-6694). This collection belongs to the section "Cancer Biomarkers".
Viewed by 31541Editor
Interests: lung cancer; molecular oncology; gene expression; cancer biomarkers; microRNA; exosomes
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
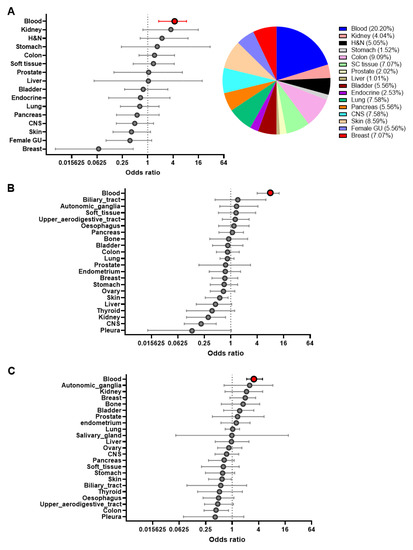
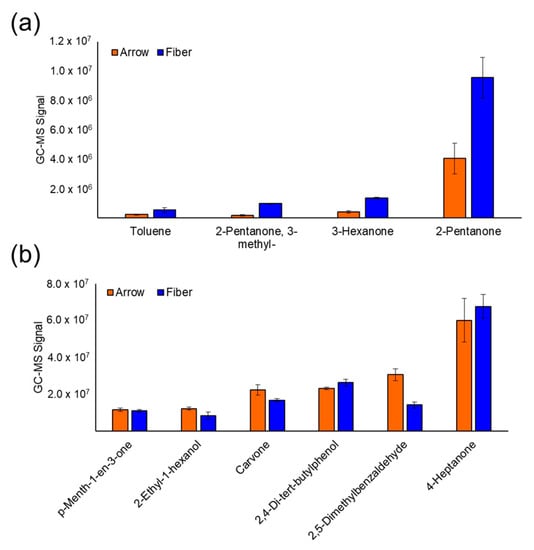
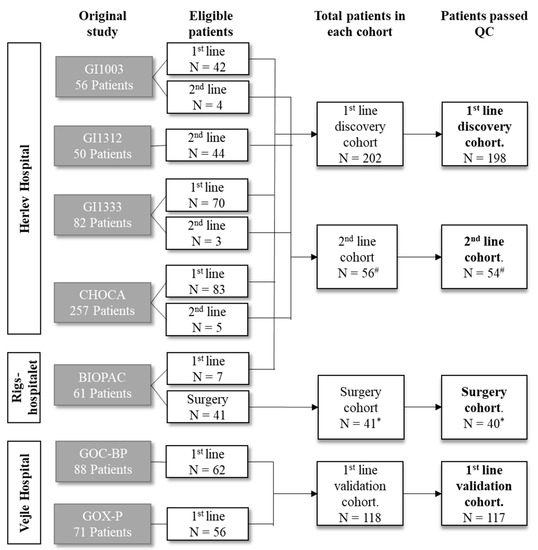
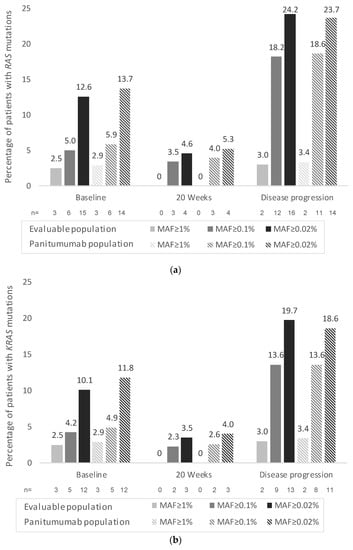
Over the next 20 years, a sharp rise in cancer cases is expected, increasing from 18.1 million people diagnosed in 2018 to an expected 29.5 million people in 2040 (Global Cancer Observatory; WHO). Cancer burden can be reduced by promoting prevention campaigns, increasing early detection, and implementing personalized cancer therapies. In such a scenario, the identification of circulating biomarkers in body fluids is emerging as a breakthrough in cancer diagnostics for the relative ease of obtaining biological samples using minimally invasive procedures before, during, and after cancer treatment; additionally, the availability of groundbreaking technologies which perform high- throughput and informative biomolecular analyses on limiting sample amounts is boosting biomarkers screening studies. Recently, several scientific publications have provided proof of principle studies which show the great advantage of using circulating biomarkers to monitor exposure to cancer risk factors, increase the accuracy of cancer screening protocols, and detect actionable therapeutic targets. Liquid biopsy shows promise for cancer screening and diagnostics, though some technical challenges still remain.
This Collection will highlight the state of the art in cancer biomarkers in body fluids and their potential application for cancer prevention/screening programs.
Dr. Fabrizio Bianchi
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cancers is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2900 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- biomarkers
- circulating biomarkers
- cancer
- early detection
- personalized therapy
- liquid biopsy