Artificial Intelligence and Machine Learning in Cancer Research
Share This Topical Collection
Editors
 Dr. Jean-Emmanuel Bibault
Dr. Jean-Emmanuel Bibault
 Dr. Jean-Emmanuel Bibault
Dr. Jean-Emmanuel Bibault
E-Mail
Website
Guest Editor
Radiation Oncology Department, Hôpital Européen Georges Pompidou, Assistance Publique - Hôpitaux de Paris, Université de Paris, F-75014 Paris, France
Interests: machine learning; artificial intelligence; prostate cancer; lung cancer; stereotactic body radiation
 Dr. Lei Xing
Dr. Lei Xing
 Dr. Lei Xing
Dr. Lei Xing
E-Mail
Website
Guest Editor
Department of Radiation Oncology, Stanford University, Stanford, CA, USA
Interests: artificial intelligence; biomedical physics; bioengineering
Topical Collection Information
Dear Colleagues,
In the near future, Artificial Intelligence and machine learning are poised to radically transform cancer care. Current research in the field of machine learning applied to oncology includes cancer screening through image analysis with deep learning, automated pathology and diagnosis, prognosis prediction and treatment personalization, drug discovery and automated treatment planning.
In this Special Issue, we invite teams working on applied AI to submit their latest and most significant research in this area. Beyond this, teams exploring interpretability and expert-augmented machine learning are invited to contribute to this new Special Issue. Studies describing new datasets and new methods are welcome. Datasets and code availability are strongly encouraged.
Dr. Jean-Emmanuel Bibault
Dr. Lei Xing
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cancers is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2900 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- Machine learning
- Artificial Intelligence
- Automation
- Prediction
- Drug discovery
Published Papers (30 papers)
Open AccessArticle
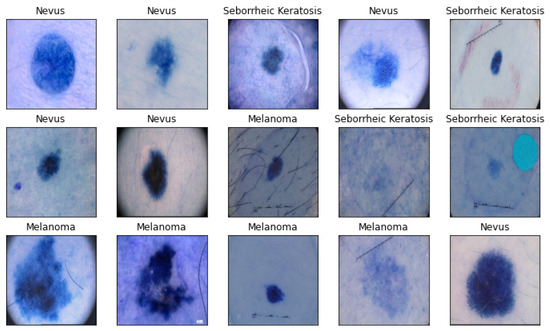
SkinLesNet: Classification of Skin Lesions and Detection of Melanoma Cancer Using a Novel Multi-Layer Deep Convolutional Neural Network
by
Muhammad Azeem, Kaveh Kiani, Taha Mansouri and Nathan Topping
Viewed by 1900
Abstract
Skin cancer is a widespread disease that typically develops on the skin due to frequent exposure to sunlight. Although cancer can appear on any part of the human body, skin cancer accounts for a significant proportion of all new cancer diagnoses worldwide. There
[...] Read more.
Skin cancer is a widespread disease that typically develops on the skin due to frequent exposure to sunlight. Although cancer can appear on any part of the human body, skin cancer accounts for a significant proportion of all new cancer diagnoses worldwide. There are substantial obstacles to the precise diagnosis and classification of skin lesions because of morphological variety and indistinguishable characteristics across skin malignancies. Recently, deep learning models have been used in the field of image-based skin-lesion diagnosis and have demonstrated diagnostic efficiency on par with that of dermatologists. To increase classification efficiency and accuracy for skin lesions, a cutting-edge multi-layer deep convolutional neural network termed SkinLesNet was built in this study. The dataset used in this study was extracted from the PAD-UFES-20 dataset and was augmented. The PAD-UFES-20-Modified dataset includes three common forms of skin lesions: seborrheic keratosis, nevus, and melanoma. To comprehensively assess SkinLesNet’s performance, its evaluation was expanded beyond the PAD-UFES-20-Modified dataset. Two additional datasets, HAM10000 and ISIC2017, were included, and SkinLesNet was compared to the widely used ResNet50 and VGG16 models. This broader evaluation confirmed SkinLesNet’s effectiveness, as it consistently outperformed both benchmarks across all datasets.
Full article
►▼
Show Figures
Open AccessArticle
Adversarial Attacks on Medical Image Classification
by
Min-Jen Tsai, Ping-Yi Lin and Ming-En Lee
Cited by 2 | Viewed by 1136
Abstract
Due to the growing number of medical images being produced by diverse radiological imaging techniques, radiography examinations with computer-aided diagnoses could greatly assist clinical applications. However, an imaging facility with just a one-pixel inaccuracy will lead to the inaccurate prediction of medical images.
[...] Read more.
Due to the growing number of medical images being produced by diverse radiological imaging techniques, radiography examinations with computer-aided diagnoses could greatly assist clinical applications. However, an imaging facility with just a one-pixel inaccuracy will lead to the inaccurate prediction of medical images. Misclassification may lead to the wrong clinical decision. This scenario is similar to the adversarial attacks on deep learning models. Therefore, one-pixel and multi-pixel level attacks on a Deep Neural Network (DNN) model trained on various medical image datasets are investigated in this study. Common multiclass and multi-label datasets are examined for one-pixel type attacks. Moreover, different experiments are conducted in order to determine how changing the number of pixels in the image may affect the classification performance and robustness of diverse DNN models. The experimental results show that it was difficult for the medical images to survive the pixel attacks, raising the issue of the accuracy of medical image classification and the importance of the model’s ability to resist these attacks for a computer-aided diagnosis.
Full article
►▼
Show Figures
Open AccessArticle
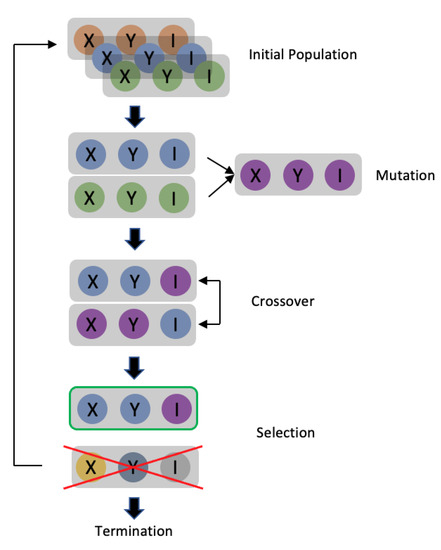
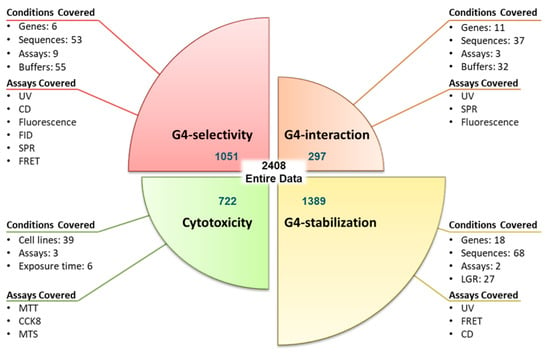
G4-QuadScreen: A Computational Tool for Identifying Multi-Target-Directed Anticancer Leads against G-Quadruplex DNA
by
Jyotsna Bhat-Ambure, Pravin Ambure, Eva Serrano-Candelas, Cristina Galiana-Roselló, Ariadna Gil-Martínez, Mario Guerrero, Margarita Martin, Jorge González-García, Enrique García-España and Rafael Gozalbes
Cited by 1 | Viewed by 1224
Abstract
The study presents ‘G4-QuadScreen’, a user-friendly computational tool for identifying MTDLs against G4s. Also, it offers a few hit MTDLs based on in silico and in vitro approaches. Multi-tasking QSAR models were developed using linear discriminant analysis and random forest machine learning techniques
[...] Read more.
The study presents ‘G4-QuadScreen’, a user-friendly computational tool for identifying MTDLs against G4s. Also, it offers a few hit MTDLs based on in silico and in vitro approaches. Multi-tasking QSAR models were developed using linear discriminant analysis and random forest machine learning techniques for predicting the responses of interest (G4 interaction, G4 stabilization, G4 selectivity, and cytotoxicity) considering the variations in the experimental conditions (e.g., G4 sequences, endpoints, cell lines, buffers, and assays). A virtual screening with G4-QuadScreen and molecular docking using YASARA (AutoDock-Vina) was performed. G4 activities were confirmed via FRET melting, FID, and cell viability assays. Validation metrics demonstrated the high discriminatory power and robustness of the models (the accuracy of all models is ~>90% for the training sets and ~>80% for the external sets). The experimental evaluations showed that ten screened MTDLs have the capacity to selectively stabilize multiple G4s. Three screened MTDLs induced a strong inhibitory effect on various human cancer cell lines. This pioneering computational study serves a tool to accelerate the search for new leads against G4s, reducing false positive outcomes in the early stages of drug discovery. The G4-QuadScreen tool is accessible on the ChemoPredictionSuite website.
Full article
►▼
Show Figures
Open AccessArticle
Automated Detection and Scoring of Tumor-Infiltrating Lymphocytes in Breast Cancer Histopathology Slides
by
Mohammad Yosofvand, Sonia Y. Khan, Rabin Dhakal, Ali Nejat, Naima Moustaid-Moussa, Rakhshanda Layeequr Rahman and Hanna Moussa
Cited by 1 | Viewed by 1271
Abstract
Detection of tumor-infiltrating lymphocytes (TILs) in cancer images has gained significant importance as these lymphocytes can be used as a biomarker in cancer detection and treatment procedures. Our goal was to develop and apply a TILs detection tool that utilizes deep learning models,
[...] Read more.
Detection of tumor-infiltrating lymphocytes (TILs) in cancer images has gained significant importance as these lymphocytes can be used as a biomarker in cancer detection and treatment procedures. Our goal was to develop and apply a TILs detection tool that utilizes deep learning models, following two sequential steps. First, based on the guidelines from the International Immuno-Oncology Biomarker Working Group (IIOBWG) on Breast Cancer, we labeled 63 large pathology imaging slides and annotated the TILs in the stroma area to create the dataset required for model development. In the second step, various machine learning models were employed and trained to detect the stroma where U-Net deep learning structure was able to achieve 98% accuracy. After detecting the stroma area, a Mask R-CNN model was employed for the TILs detection task. The R-CNN model detected the TILs in various images and was used as the backbone analysis network for the GUI development of the TILs detection tool. This is the first study to combine two deep learning models for TILs detection at the cellular level in breast tumor histopathology slides. Our novel approach can be applied to scoring TILs in large cancer slides. Statistical analysis showed that the output of the implemented approach had 95% concordance with the scores assigned by the pathologists, with a
p-value of 0.045 (n = 63). This demonstrated that the results from the developed software were statistically meaningful and highly accurate. The implemented approach in analyzing whole tumor histology slides and the newly developed TILs detection tool can be used for research purposes in biomedical and pathology applications and it can provide researchers and clinicians with the TIL score for various input images. Future research using additional breast cancer slides from various sources for further training and validation of the developed models is necessary for more inclusive, rigorous, and robust clinical applications.
Full article
►▼
Show Figures
Open AccessArticle
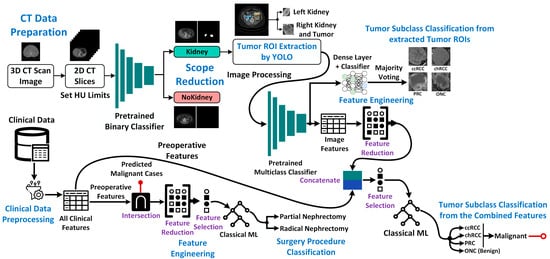
Kidney Cancer Diagnosis and Surgery Selection by Machine Learning from CT Scans Combined with Clinical Metadata
by
Sakib Mahmud, Tariq O. Abbas, Adam Mushtak, Johayra Prithula and Muhammad E. H. Chowdhury
Cited by 6 | Viewed by 2845
Abstract
Kidney cancers are one of the most common malignancies worldwide. Accurate diagnosis is a critical step in the management of kidney cancer patients and is influenced by multiple factors including tumor size or volume, cancer types and stages, etc. For malignant tumors, partial
[...] Read more.
Kidney cancers are one of the most common malignancies worldwide. Accurate diagnosis is a critical step in the management of kidney cancer patients and is influenced by multiple factors including tumor size or volume, cancer types and stages, etc. For malignant tumors, partial or radical surgery of the kidney might be required, but for clinicians, the basis for making this decision is often unclear. Partial nephrectomy could result in patient death due to cancer if kidney removal was necessary, whereas radical nephrectomy in less severe cases could resign patients to lifelong dialysis or need for future transplantation without sufficient cause. Using machine learning to consider clinical data alongside computed tomography images could potentially help resolve some of these surgical ambiguities, by enabling a more robust classification of kidney cancers and selection of optimal surgical approaches. In this study, we used the publicly available KiTS dataset of contrast-enhanced CT images and corresponding patient metadata to differentiate four major classes of kidney cancer: clear cell (ccRCC), chromophobe (chRCC), papillary (pRCC) renal cell carcinoma, and oncocytoma (ONC). We rationalized these data to overcome the high field of view (FoV), extract tumor regions of interest (ROIs), classify patients using deep machine-learning models, and extract/post-process CT image features for combination with clinical data. Regardless of marked data imbalance, our combined approach achieved a high level of performance (85.66% accuracy, 84.18% precision, 85.66% recall, and 84.92% F1-score). When selecting surgical procedures for malignant tumors (RCC), our method proved even more reliable (90.63% accuracy, 90.83% precision, 90.61% recall, and 90.50% F1-score). Using feature ranking, we confirmed that tumor volume and cancer stage are the most relevant clinical features for predicting surgical procedures. Once fully mature, the approach we propose could be used to assist surgeons in performing nephrectomies by guiding the choices of optimal procedures in individual patients with kidney cancer.
Full article
►▼
Show Figures
Open AccessArticle
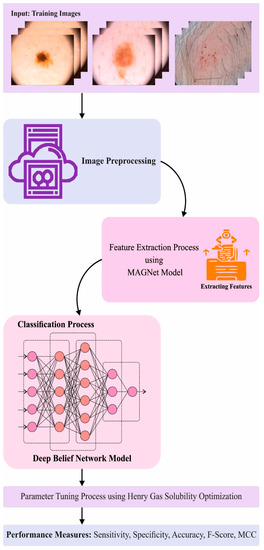
Henry Gas Solubility Optimization Algorithm based Feature Extraction in Dermoscopic Images Analysis of Skin Cancer
by
Marwa Obayya, Adeeb Alhebri, Mashael Maashi, Ahmed S. Salama, Anwer Mustafa Hilal, Mohamed Ibrahim Alsaid, Azza Elneil Osman and Amani A. Alneil
Cited by 6 | Viewed by 1813
Abstract
Artificial Intelligence (AI) techniques have changed the general perceptions about medical diagnostics, especially after the introduction and development of Convolutional Neural Networks (CNN) and advanced Deep Learning (DL) and Machine Learning (ML) approaches. In general, dermatologists visually inspect the images and assess the
[...] Read more.
Artificial Intelligence (AI) techniques have changed the general perceptions about medical diagnostics, especially after the introduction and development of Convolutional Neural Networks (CNN) and advanced Deep Learning (DL) and Machine Learning (ML) approaches. In general, dermatologists visually inspect the images and assess the morphological variables such as borders, colors, and shapes to diagnose the disease. In this background, AI techniques make use of algorithms and computer systems to mimic the cognitive functions of the human brain and assist clinicians and researchers. In recent years, AI has been applied extensively in the domain of dermatology, especially for the detection and classification of skin cancer and other general skin diseases. In this research article, the authors propose an Optimal Multi-Attention Fusion Convolutional Neural Network-based Skin Cancer Diagnosis (MAFCNN-SCD) technique for the detection of skin cancer in dermoscopic images. The primary aim of the proposed MAFCNN-SCD technique is to classify skin cancer on dermoscopic images. In the presented MAFCNN-SCD technique, the data pre-processing is performed at the initial stage. Next, the MAFNet method is applied as a feature extractor with Henry Gas Solubility Optimization (HGSO) algorithm as a hyperparameter optimizer. Finally, the Deep Belief Network (DBN) method is exploited for the detection and classification of skin cancer. A sequence of simulations was conducted to establish the superior performance of the proposed MAFCNN-SCD approach. The comprehensive comparative analysis outcomes confirmed the supreme performance of the proposed MAFCNN-SCD technique over other methodologies.
Full article
►▼
Show Figures
Open AccessArticle
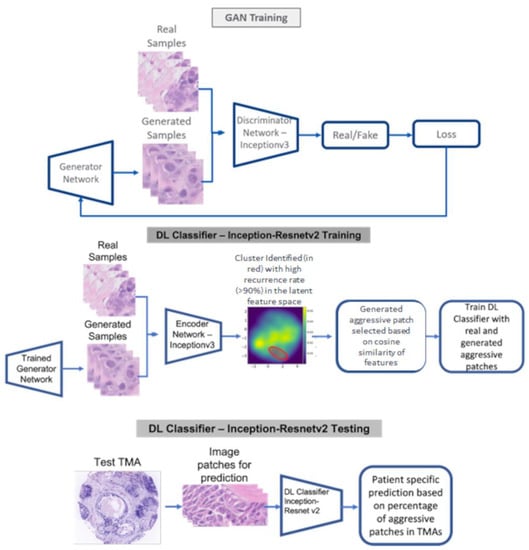
Predicting Breast Cancer Events in Ductal Carcinoma In Situ (DCIS) Using Generative Adversarial Network Augmented Deep Learning Model
by
Soumya Ghose, Sanghee Cho, Fiona Ginty, Elizabeth McDonough, Cynthia Davis, Zhanpan Zhang, Jhimli Mitra, Adrian L. Harris, Aye Aye Thike, Puay Hoon Tan, Yesim Gökmen-Polar and Sunil S. Badve
Cited by 1 | Viewed by 1515
Abstract
Standard clinicopathological parameters (age, growth pattern, tumor size, margin status, and grade) have been shown to have limited value in predicting recurrence in ductal carcinoma in situ (DCIS) patients. Early and accurate recurrence prediction would facilitate a more aggressive treatment policy for high-risk
[...] Read more.
Standard clinicopathological parameters (age, growth pattern, tumor size, margin status, and grade) have been shown to have limited value in predicting recurrence in ductal carcinoma in situ (DCIS) patients. Early and accurate recurrence prediction would facilitate a more aggressive treatment policy for high-risk patients (mastectomy or adjuvant radiation therapy), and simultaneously reduce over-treatment of low-risk patients. Generative adversarial networks (GAN) are a class of DL models in which two adversarial neural networks, generator and discriminator, compete with each other to generate high quality images. In this work, we have developed a deep learning (DL) classification network that predicts breast cancer events (BCEs) in DCIS patients using hematoxylin and eosin (H & E) images. The DL classification model was trained on 67 patients using image patches from the actual DCIS cores and GAN generated image patches to predict breast cancer events (BCEs). The hold-out validation dataset (
n = 66) had an AUC of 0.82. Bayesian analysis further confirmed the independence of the model from classical clinicopathological parameters. DL models of H & E images may be used as a risk stratification strategy for DCIS patients to personalize therapy.
Full article
►▼
Show Figures
Open AccessArticle
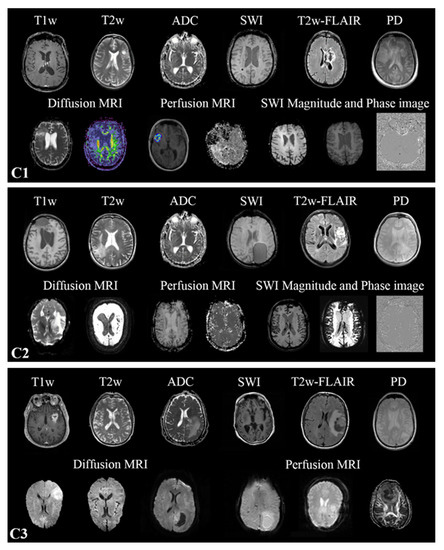
MR-Class: A Python Tool for Brain MR Image Classification Utilizing One-vs-All DCNNs to Deal with the Open-Set Recognition Problem
by
Patrick Salome, Francesco Sforazzini, Gianluca Brugnara, Andreas Kudak, Matthias Dostal, Christel Herold-Mende, Sabine Heiland, Jürgen Debus, Amir Abdollahi and Maximilian Knoll
Viewed by 2677
Abstract
Background: MR image classification in datasets collected from multiple sources is complicated by inconsistent and missing DICOM metadata. Therefore, we aimed to establish a method for the efficient automatic classification of MR brain sequences. Methods: Deep convolutional neural networks (DCNN) were trained as
[...] Read more.
Background: MR image classification in datasets collected from multiple sources is complicated by inconsistent and missing DICOM metadata. Therefore, we aimed to establish a method for the efficient automatic classification of MR brain sequences. Methods: Deep convolutional neural networks (DCNN) were trained as one-vs-all classifiers to differentiate between six classes: T1 weighted (w), contrast-enhanced T1w, T2w, T2w-FLAIR, ADC, and SWI. Each classifier yields a probability, allowing threshold-based and relative probability assignment while excluding images with low probability (label: unknown, open-set recognition problem). Data from three high-grade glioma (HGG) cohorts was assessed; C1 (320 patients, 20,101 MRI images) was used for training, while C2 (197, 11,333) and C3 (256, 3522) were for testing. Two raters manually checked images through an interactive labeling tool. Finally, MR-Class’ added value was evaluated via radiomics model performance for progression-free survival (PFS) prediction in C2, utilizing the concordance index (C-I). Results: Approximately 10% of annotation errors were observed in each cohort between the DICOM series descriptions and the derived labels. MR-Class accuracy was 96.7% [95% Cl: 95.8, 97.3] for C2 and 94.4% [93.6, 96.1] for C3. A total of 620 images were misclassified; manual assessment of those frequently showed motion artifacts or alterations of anatomy by large tumors. Implementation of MR-Class increased the PFS model C-I by 14.6% on average, compared to a model trained without MR-Class. Conclusions: We provide a DCNN-based method for the sequence classification of brain MR images and demonstrate its usability in two independent HGG datasets.
Full article
►▼
Show Figures
Open AccessArticle
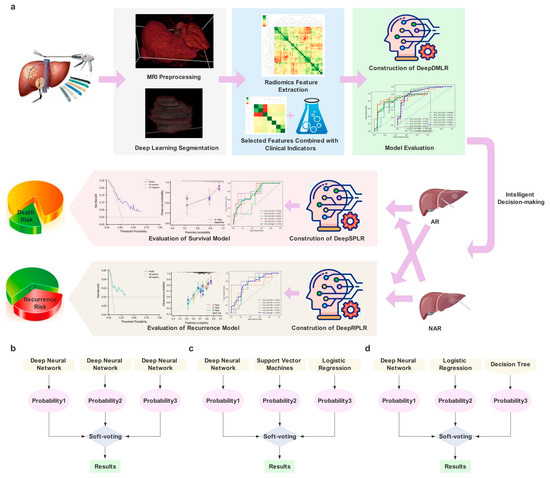
Data-Driven Assisted Decision Making for Surgical Procedure of Hepatocellular Carcinoma Resection and Prognostic Prediction: Development and Validation of Machine Learning Models
by
Liyang Wang, Danjun Song, Wentao Wang, Chengquan Li, Yiming Zhou, Jiaping Zheng, Shengxiang Rao, Xiaoying Wang, Guoliang Shao, Jiabin Cai, Shizhong Yang and Jiahong Dong
Cited by 1 | Viewed by 1498
Abstract
Background
: Currently, surgical decisions for hepatocellular carcinoma (HCC) resection are difficult and not sufficiently personalized. We aimed to develop and validate data driven prediction models to assist surgeons in selecting the optimal surgical procedure for patients. Methods
: Retrospective data from 361
[...] Read more.
Background
: Currently, surgical decisions for hepatocellular carcinoma (HCC) resection are difficult and not sufficiently personalized. We aimed to develop and validate data driven prediction models to assist surgeons in selecting the optimal surgical procedure for patients. Methods
: Retrospective data from 361 HCC patients who underwent radical resection in two institutions were included. End-to-end deep learning models were built to automatically segment lesions from the arterial phase (AP) of preoperative dynamic contrast enhanced magnetic resonance imaging (DCE-MRI). Clinical baseline characteristics and radiomic features were rigorously screened. The effectiveness of radiomic features and radiomic-clinical features was also compared. Three ensemble learning models were proposed to perform the surgical procedure decision and the overall survival (OS) and recurrence-free survival (RFS) predictions after taking different solutions, respectively. Results
: SegFormer performed best in terms of automatic segmentation, achieving a Mean Intersection over Union (mIoU) of 0.8860. The five-fold cross-validation results showed that inputting radiomic-clinical features outperformed using only radiomic features. The proposed models all outperformed the other mainstream ensemble models. On the external test set, the area under the receiver operating characteristic curve (AUC) of the proposed decision model was 0.7731, and the performance of the prognostic prediction models was also relatively excellent. The application web server based on automatic lesion segmentation was deployed and is available online. Conclusions
: In this study, we developed and externally validated the surgical decision-making procedures and prognostic prediction models for HCC for the first time, and the results demonstrated relatively accurate predictions and strong generalizations, which are expected to help clinicians optimize surgical procedures.
Full article
►▼
Show Figures
Open AccessArticle
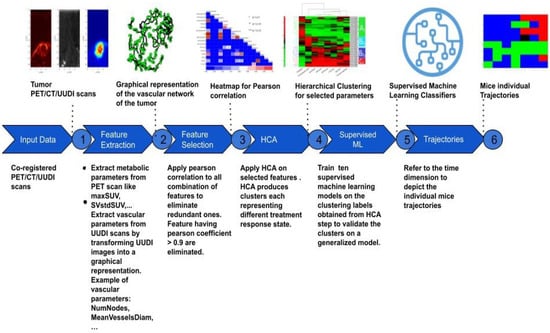
Machine Learning of Multi-Modal Tumor Imaging Reveals Trajectories of Response to Precision Treatment
by
Nesrin Mansouri, Daniel Balvay, Omar Zenteno, Caterina Facchin, Thulaciga Yoganathan, Thomas Viel, Joaquin Lopez Herraiz, Bertrand Tavitian and Mailyn Pérez-Liva
Cited by 2 | Viewed by 2113
Abstract
The standard assessment of response to cancer treatments is based on gross tumor characteristics, such as tumor size or glycolysis, which provide very indirect information about the effect of precision treatments on the pharmacological targets of tumors. Several advanced imaging modalities allow for
[...] Read more.
The standard assessment of response to cancer treatments is based on gross tumor characteristics, such as tumor size or glycolysis, which provide very indirect information about the effect of precision treatments on the pharmacological targets of tumors. Several advanced imaging modalities allow for the visualization of targeted tumor hallmarks. Descriptors extracted from these images can help establishing new classifications of precision treatment response. We propose a machine learning (ML) framework to analyze metabolic–anatomical–vascular imaging features from positron emission tomography, ultrafast Doppler, and computed tomography in a mouse model of paraganglioma undergoing anti-angiogenic treatment with sunitinib. Imaging features from the follow-up of sunitinib-treated (
n = 8, imaged once-per-week/6-weeks) and sham-treated (
n = 8, imaged once-per-week/3-weeks) mice groups were dimensionally reduced and analyzed with hierarchical clustering Analysis (HCA). The classes extracted from HCA were used with 10 ML classifiers to find a generalized tumor stage prediction model, which was validated with an independent dataset of sunitinib-treated mice. HCA provided three stages of treatment response that were validated using the best-performing ML classifier. The Gaussian naive Bayes classifier showed the best performance, with a training accuracy of 98.7 and an average area under curve of 100. Our results show that metabolic–anatomical–vascular markers allow defining treatment response trajectories that reflect the efficacy of an anti-angiogenic drug on the tumor target hallmark.
Full article
►▼
Show Figures
Open AccessArticle
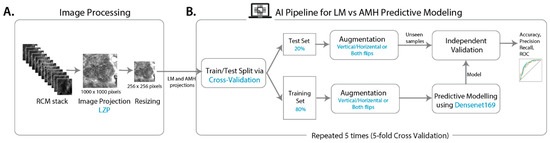
Computer-Aided Diagnosis of Melanoma Subtypes Using Reflectance Confocal Images
by
Ankita Mandal, Siddhaant Priyam, Hsien Herbert Chan, Bruna Melhoranse Gouveia, Pascale Guitera, Yang Song, Matthew Arthur Barrington Baker and Fatemeh Vafaee
Cited by 2 | Viewed by 1723
Abstract
Lentigo maligna (LM) is an early form of pre-invasive melanoma that predominantly affects sun-exposed areas such as the face. LM is highly treatable when identified early but has an ill-defined clinical border and a high rate of recurrence. Atypical intraepidermal melanocytic proliferation (AIMP),
[...] Read more.
Lentigo maligna (LM) is an early form of pre-invasive melanoma that predominantly affects sun-exposed areas such as the face. LM is highly treatable when identified early but has an ill-defined clinical border and a high rate of recurrence. Atypical intraepidermal melanocytic proliferation (AIMP), also known as atypical melanocytic hyperplasia (AMH), is a histological description that indicates melanocytic proliferation with uncertain malignant potential. Clinically and histologically, AIMP can be difficult to distinguish from LM, and indeed AIMP may, in some cases, progress to LM. The early diagnosis and distinction of LM from AIMP are important since LM requires a definitive treatment. Reflectance confocal microscopy (RCM) is an imaging technique often used to investigate these lesions non-invasively, without biopsy. However, RCM equipment is often not readily available, nor is the associated expertise for RCM image interpretation easy to find. Here, we implemented a machine learning classifier using popular convolutional neural network (CNN) architectures and demonstrated that it could correctly classify lesions between LM and AIMP on biopsy-confirmed RCM image stacks. We identified local z-projection (LZP) as a recent fast approach for projecting a 3D image into 2D while preserving information and achieved high-accuracy machine classification with minimal computational requirements.
Full article
►▼
Show Figures
Open AccessArticle
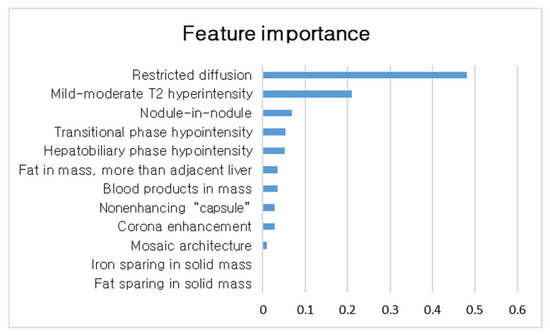
Utilization of a Machine Learning Algorithm for the Application of Ancillary Features to LI-RADS Categories LR3 and LR4 on Gadoxetate Disodium-Enhanced MRI
by
Seongkeun Park, Jieun Byun and Sook Min Hwang
Cited by 2 | Viewed by 1164
Abstract
Background: This study aimed to identify the important ancillary features (AFs) and determine the utilization of a machine-learning-based strategy for applying AFs for LI-RADS LR3/4 observations on gadoxetate disodium-enhanced MRI. Methods: We retrospectively analyzed MRI features of LR3/4 determined with only major features.
[...] Read more.
Background: This study aimed to identify the important ancillary features (AFs) and determine the utilization of a machine-learning-based strategy for applying AFs for LI-RADS LR3/4 observations on gadoxetate disodium-enhanced MRI. Methods: We retrospectively analyzed MRI features of LR3/4 determined with only major features. Uni- and multivariate analyses and random forest analysis were performed to identify AFs associated with HCC. A decision tree algorithm of applying AFs for LR3/4 was compared with other alternative strategies using McNemar’s test. Results: We evaluated 246 observations from 165 patients. In multivariate analysis, restricted diffusion and mild–moderate T2 hyperintensity showed independent associations with HCC (odds ratios: 12.4 [
p < 0.001] and 2.5 [
p = 0.02]). In random forest analysis, restricted diffusion is the most important feature for HCC. Our decision tree algorithm showed higher AUC, sensitivity, and accuracy (0.84, 92.0%, and 84.5%) than the criteria of usage of restricted diffusion (0.78, 64.5%, and 76.4%; all
p < 0.05); however, our decision tree algorithm showed lower specificity than the criterion of usage of restricted diffusion (71.1% vs. 91.3%;
p < 0.001). Conclusion: Our decision tree algorithm of applying AFs for LR3/4 shows significantly increased AUC, sensitivity, and accuracy but reduced specificity. These appear to be more appropriate in certain circumstances in which there is an emphasis on the early detection of HCC.
Full article
►▼
Show Figures
Open AccessArticle
Improving the Post-Operative Prediction of BCR-Free Survival Time with mRNA Variables and Machine Learning
by
Autumn O’Donnell, Eric Wolsztynski, Michael Cronin and Shirin Moghaddam
Cited by 2 | Viewed by 1825
Abstract
Predicting the risk of, and time to biochemical recurrence (BCR) in prostate cancer patients post-operatively is critical in patient treatment decision pathways following surgical intervention. This study aimed to investigate the predictive potential of mRNA information to improve upon reference nomograms and clinical-only
[...] Read more.
Predicting the risk of, and time to biochemical recurrence (BCR) in prostate cancer patients post-operatively is critical in patient treatment decision pathways following surgical intervention. This study aimed to investigate the predictive potential of mRNA information to improve upon reference nomograms and clinical-only models, using a dataset of 187 patients that includes over 20,000 features. Several machine learning methodologies were implemented for the analysis of censored patient follow-up information with such high-dimensional genomic data. Our findings demonstrated the potential of inclusion of mRNA information for BCR-free survival prediction. A random survival forest pipeline was found to achieve high predictive performance with respect to discrimination, calibration, and net benefit. Two mRNA variables, namely ESM1 and DHAH8, were identified as consistently strong predictors with this dataset.
Full article
►▼
Show Figures
Open AccessArticle
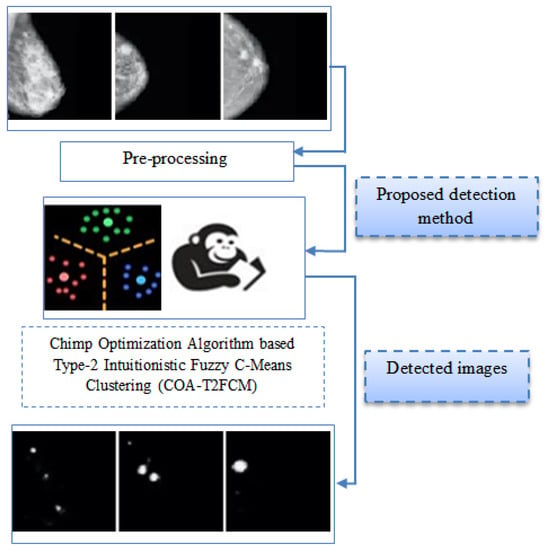
Chimp Optimization Algorithm Influenced Type-2 Intuitionistic Fuzzy C-Means Clustering-Based Breast Cancer Detection System
by
Prasanalakshmi Balaji, Vasanthi Muniasamy, Syeda Meraj Bilfaqih, Anandhavalli Muniasamy, Sridevi Tharanidharan, Devi Mani and Linda Elzubir Gasm Alsid
Cited by 4 | Viewed by 3031
Abstract
In recent years, breast cancer detection is an important area of concentration towards curative image dispensation and exploration. Detection of a disease at an early stage is an important factor in taking it to the next level of treatment. Accuracy plays an important
[...] Read more.
In recent years, breast cancer detection is an important area of concentration towards curative image dispensation and exploration. Detection of a disease at an early stage is an important factor in taking it to the next level of treatment. Accuracy plays an important role in the detection of disease. COA-T2FCM (Chimp Optimization Algorithm Based Type-2 Intuitionistic Fuzzy C-Means Clustering) is constructed for detection of such malignancy with the highest accuracy in this paper. The proposed detection process is designed with the combination of type-2 intuitionistic fuzzy c-means clustering in addition to oppositional function. In the type-2 intuitionistic fuzzy c-means clustering, the efficient cluster center can be preferred using the chimp optimization algorithm. Initially, the objective function of the type-2 intuitionistic fuzzy c-means clustering is considered. The chimp optimization algorithm is utilized to optimize the cluster center and fuzzifier in the clustering method. The projected technique is implemented, and in addition, performance metrics such as specificity, sensitivity, accuracy, Jaccard Similarity Index (JSI), and Dice Similarity Coefficient (DSC) are assessed. The projected technique is compared with the conventional technique such as fuzzy c means clustering and k mean clustering methods. The resulting method was also compared with existing methods to ensure the accuracy in the proposed method. The proposed algorithm is tested for its effectiveness on the mammogram images of the three different datasets collected from the Mini–Mammographic Image Analysis Society (Mini–MIAS), the Digital Database for Screening Mammography (DDSM), and Inbreast. The accuracy and Jaccard index score are generally used to measure the similarity between the proposed output and the actual cancer affected regions from the image considered. On an average the proposed method achieved an accuracy of 97.29% and JSI of 95%
Full article
►▼
Show Figures
Open AccessArticle
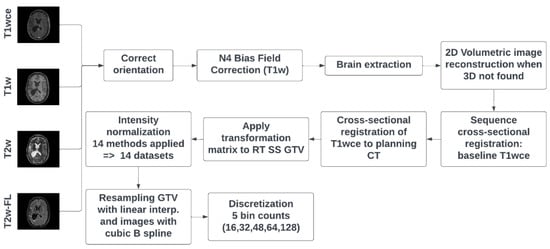
MR Intensity Normalization Methods Impact Sequence Specific Radiomics Prognostic Model Performance in Primary and Recurrent High-Grade Glioma
by
Patrick Salome, Francesco Sforazzini, Gianluca Brugnara, Andreas Kudak, Matthias Dostal, Christel Herold-Mende, Sabine Heiland, Jürgen Debus, Amir Abdollahi and Maximilian Knoll
Cited by 3 | Viewed by 2270
Abstract
Purpose: This study investigates the impact of different intensity normalization (IN) methods on the overall survival (OS) radiomics models’ performance of MR sequences in primary (pHGG) and recurrent high-grade glioma (rHGG). Methods: MR scans acquired before radiotherapy were retrieved from two independent cohorts
[...] Read more.
Purpose: This study investigates the impact of different intensity normalization (IN) methods on the overall survival (OS) radiomics models’ performance of MR sequences in primary (pHGG) and recurrent high-grade glioma (rHGG). Methods: MR scans acquired before radiotherapy were retrieved from two independent cohorts (rHGG C1: 197, pHGG C2: 141) from multiple scanners (15, 14). The sequences are T1 weighted (w), contrast-enhanced T1w (T1wce), T2w, and T2w-FLAIR. Sequence-specific significant features (SF) associated with OS, extracted from the tumour volume, were derived after applying 15 different IN methods. Survival analyses were conducted using Cox proportional hazard (CPH) and Poisson regression (POI) models. A ranking score was assigned based on the 10-fold cross-validated (CV) concordance index (C-I), mean square error (MSE), and the Akaike information criterion (AICs), to evaluate the methods’ performance. Results: Scatter plots of the 10-CV C-I and MSE against the AIC showed an impact on the survival predictions between the IN methods and MR sequences (C1/C2 C-I range: 0.62–0.71/0.61–0.72, MSE range: 0.20–0.42/0.13–0.22). White stripe showed stable results for T1wce (C1/C2 C-I: 0.71/0.65, MSE: 0.21/0.14). Combat (0.68/0.62, 0.22/0.15) and histogram matching (HM, 0.67/0.64, 0.22/0.15) showed consistent prediction results for T2w models. They were also the top-performing methods for T1w in C2 (Combat: 0.67, 0.13; HM: 0.67, 0.13); however, only HM achieved high predictions in C1 (0.66, 0.22). After eliminating IN impacted SF using Spearman’s rank-order correlation coefficient, a mean decrease in the C-I and MSE of 0.05 and 0.03 was observed in all four sequences. Conclusion: The IN method impacted the predictive power of survival models; thus, performance is sequence-dependent.
Full article
►▼
Show Figures
Open AccessArticle
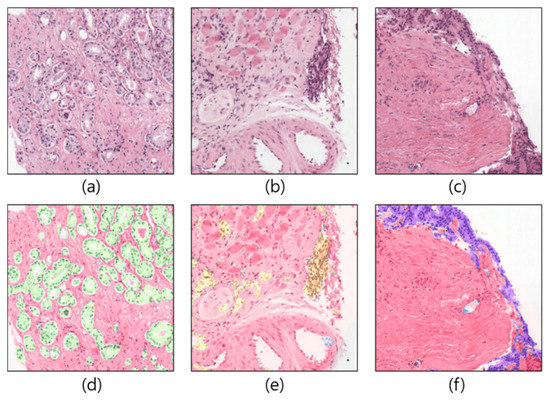
Region Segmentation of Whole-Slide Images for Analyzing Histological Differentiation of Prostate Adenocarcinoma Using Ensemble EfficientNetB2 U-Net with Transfer Learning Mechanism
by
Kobiljon Ikromjanov, Subrata Bhattacharjee, Rashadul Islam Sumon, Yeong-Byn Hwang, Hafizur Rahman, Myung-Jae Lee, Hee-Cheol Kim, Eunhyang Park, Nam-Hoon Cho and Heung-Kook Choi
Cited by 3 | Viewed by 2314
Abstract
Recent advances in computer-aided detection via deep learning (DL) now allow for prostate cancer to be detected automatically and recognized with extremely high accuracy, much like other medical diagnoses and prognoses. However, researchers are still limited by the Gleason scoring system. The histopathological
[...] Read more.
Recent advances in computer-aided detection via deep learning (DL) now allow for prostate cancer to be detected automatically and recognized with extremely high accuracy, much like other medical diagnoses and prognoses. However, researchers are still limited by the Gleason scoring system. The histopathological analysis involved in assigning the appropriate score is a rigorous, time-consuming manual process that is constrained by the quality of the material and the pathologist’s level of expertise. In this research, we implemented a DL model using transfer learning on a set of histopathological images to segment cancerous and noncancerous areas in whole-slide images (WSIs). In this approach, the proposed Ensemble U-net model was applied for the segmentation of stroma, cancerous, and benign areas. The WSI dataset of prostate cancer was collected from the Kaggle repository, which is publicly available online. A total of 1000 WSIs were used for region segmentation. From this, 8100 patch images were used for training, and 900 for testing. The proposed model demonstrated an average dice coefficient (DC), intersection over union (IoU), and Hausdorff distance of 0.891, 0.811, and 15.9, respectively, on the test set, with corresponding masks of patch images. The manipulation of the proposed segmentation model improves the ability of the pathologist to predict disease outcomes, thus enhancing treatment efficacy by isolating the cancerous regions in WSIs.
Full article
►▼
Show Figures
Open AccessArticle
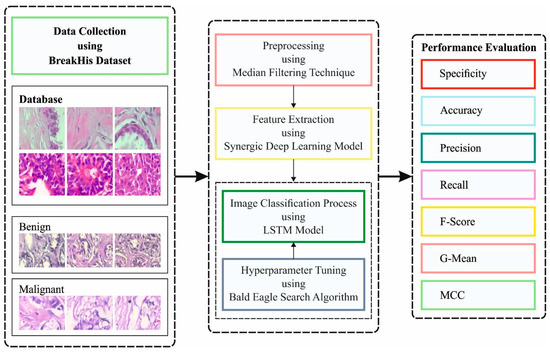
Improved Bald Eagle Search Optimization with Synergic Deep Learning-Based Classification on Breast Cancer Imaging
by
Manar Ahmed Hamza, Hanan Abdullah Mengash, Mohamed K Nour, Naif Alasmari, Amira Sayed A. Aziz, Gouse Pasha Mohammed, Abu Sarwar Zamani and Amgad Atta Abdelmageed
Cited by 3 | Viewed by 1271
Abstract
Medical imaging has attracted growing interest in the field of healthcare regarding breast cancer (BC). Globally, BC is a major cause of mortality amongst women. Now, the examination of histopathology images is the medical gold standard for cancer diagnoses. However, the manual process
[...] Read more.
Medical imaging has attracted growing interest in the field of healthcare regarding breast cancer (BC). Globally, BC is a major cause of mortality amongst women. Now, the examination of histopathology images is the medical gold standard for cancer diagnoses. However, the manual process of microscopic inspections is a laborious task, and the results might be misleading as a result of human error occurring. Thus, the computer-aided diagnoses (CAD) system can be utilized for accurately detecting cancer within essential time constraints, as earlier diagnosis is the key to curing cancer. The classification and diagnosis of BC utilizing the deep learning algorithm has gained considerable attention. This article presents a model of an improved bald eagle search optimization with a synergic deep learning mechanism for breast cancer diagnoses using histopathological images (IBESSDL-BCHI). The proposed IBESSDL-BCHI model concentrates on the identification and classification of BC using HIs. To do so, the presented IBESSDL-BCHI model follows an image preprocessing method using a median filtering (MF) technique as a preprocessing step. In addition, feature extraction using a synergic deep learning (SDL) model is carried out, and the hyperparameters related to the SDL mechanism are tuned by the use of the IBES model. Lastly, long short-term memory (LSTM) was utilized to precisely categorize the HIs into two major classes, such as benign and malignant. The performance validation of the IBESSDL-BCHI system was tested utilizing the benchmark dataset, and the results demonstrate that the IBESSDL-BCHI model has shown better general efficiency for BC classification.
Full article
►▼
Show Figures
Open AccessArticle
Design of a Honey Badger Optimization Algorithm with a Deep Transfer Learning-Based Osteosarcoma Classification Model
by
Thavavel Vaiyapuri, Akshya Jothi, Kanagaraj Narayanasamy, Kartheeban Kamatchi, Seifedine Kadry and Jungeun Kim
Cited by 5 | Viewed by 2007
Abstract
Osteosarcoma is one of the aggressive bone tumors with numerous histologic patterns. Histopathological inspection is a crucial criterion in the medical diagnosis of Osteosarcoma. Due to the advancement of computing power and hardware technology, pathological image analysis system based on artificial intelligence (AI)
[...] Read more.
Osteosarcoma is one of the aggressive bone tumors with numerous histologic patterns. Histopathological inspection is a crucial criterion in the medical diagnosis of Osteosarcoma. Due to the advancement of computing power and hardware technology, pathological image analysis system based on artificial intelligence (AI) were more commonly used. But classifying many intricate pathology images by hand will be challenging for pathologists. The lack of labeling data makes the system difficult to build and costly. This article designs a Honey Badger Optimization with Deep Learning based Automated Osteosarcoma Classification (HBODL-AOC) model. The HBODL-AOC technique’s goal is to identify osteosarcoma’s existence using medical images. In the presented HBODL-AOC technique, image preprocessing is initially performed by contrast enhancement technique. For feature extraction, the HBODL-AOC technique employs a deep convolutional neural network-based Mobile networks (MobileNet) model with an Adam optimizer for hyperparameter tuning. Finally, the adaptive neuro-fuzzy inference system (ANFIS) approach is implemented for the HBO (Honey Badger Optimization) algorithm can tune osteosarcoma classification and the membership function (MF). To demonstrate the enhanced classification performance of the HBODL-AOC approach, a sequence of simulations was performed. The extensive simulation analysis portrayed the improved performance of the HBODL-AOC technique over existing DL models.
Full article
►▼
Show Figures
Open AccessArticle
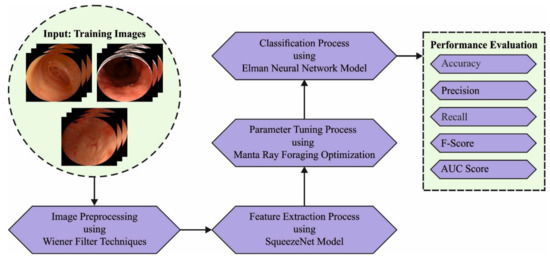
Manta Ray Foraging Optimization Transfer Learning-Based Gastric Cancer Diagnosis and Classification on Endoscopic Images
by
Fadwa Alrowais, Saud S. Alotaibi, Radwa Marzouk, Ahmed S. Salama, Mohammed Rizwanullah, Abu Sarwar Zamani, Amgad Atta Abdelmageed and Mohamed I. Eldesouki
Cited by 2 | Viewed by 2615
Abstract
Gastric cancer (GC) diagnoses using endoscopic images have gained significant attention in the healthcare sector. The recent advancements of computer vision (CV) and deep learning (DL) technologies pave the way for the design of automated GC diagnosis models. Therefore, this study develops a
[...] Read more.
Gastric cancer (GC) diagnoses using endoscopic images have gained significant attention in the healthcare sector. The recent advancements of computer vision (CV) and deep learning (DL) technologies pave the way for the design of automated GC diagnosis models. Therefore, this study develops a new Manta Ray Foraging Optimization Transfer Learning technique that is based on Gastric Cancer Diagnosis and Classification (MRFOTL-GCDC) using endoscopic images. For enhancing the quality of the endoscopic images, the presented MRFOTL-GCDC technique executes the Wiener filter (WF) to perform a noise removal process. In the presented MRFOTL-GCDC technique, MRFO with SqueezeNet model is used to derive the feature vectors. Since the trial-and-error hyperparameter tuning is a tedious process, the MRFO algorithm-based hyperparameter tuning results in enhanced classification results. Finally, the Elman Neural Network (ENN) model is utilized for the GC classification. To depict the enhanced performance of the presented MRFOTL-GCDC technique, a widespread simulation analysis is executed. The comparison study reported the improvement of the MRFOTL-GCDC technique for endoscopic image classification purposes with an improved accuracy of 99.25%.
Full article
►▼
Show Figures
Open AccessArticle
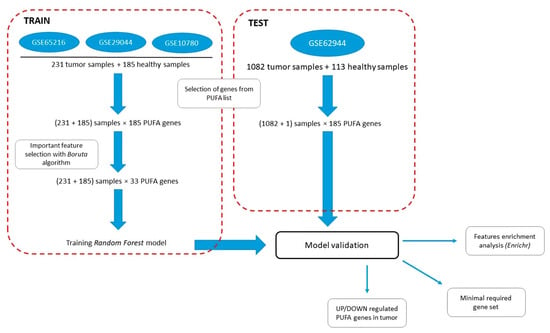
Investigation of the Role of PUFA Metabolism in Breast Cancer Using a Rank-Based Random Forest Algorithm
by
Mariia V. Guryleva, Dmitry D. Penzar, Dmitry V. Chistyakov, Andrey A. Mironov, Alexander V. Favorov and Marina G. Sergeeva
Cited by 2 | Viewed by 1762
Abstract
Polyunsaturated fatty acid (PUFA) metabolism is currently a focus in cancer research due to PUFAs functioning as structural components of the membrane matrix, as fuel sources for energy production, and as sources of secondary messengers, so called oxylipins, important players of inflammatory processes.
[...] Read more.
Polyunsaturated fatty acid (PUFA) metabolism is currently a focus in cancer research due to PUFAs functioning as structural components of the membrane matrix, as fuel sources for energy production, and as sources of secondary messengers, so called oxylipins, important players of inflammatory processes. Although breast cancer (BC) is the leading cause of cancer death among women worldwide, no systematic study of PUFA metabolism as a system of interrelated processes in this disease has been carried out. Here, we implemented a Boruta-based feature selection algorithm to determine the list of most important PUFA metabolism genes altered in breast cancer tissues compared with in normal tissues. A rank-based Random Forest (RF) model was built on the selected gene list (33 genes) and applied to predict the cancer phenotype to ascertain the PUFA genes involved in cancerogenesis. It showed high-performance of dichotomic classification (balanced accuracy of 0.94, ROC AUC 0.99) We also retrieved a list of the important PUFA genes (46 genes) that differed between molecular subtypes at the level of breast cancer molecular subtypes. The balanced accuracy of the classification model built on the specified genes was 0.82, while the ROC AUC for the sensitivity analysis was 0.85. Specific patterns of PUFA metabolic changes were obtained for each molecular subtype of breast cancer. These results show evidence that (1) PUFA metabolism genes are critical for the pathogenesis of breast cancer; (2) BC subtypes differ in PUFA metabolism genes expression; and (3) the lists of genes selected in the models are enriched with genes involved in the metabolism of signaling lipids.
Full article
►▼
Show Figures
Open AccessArticle
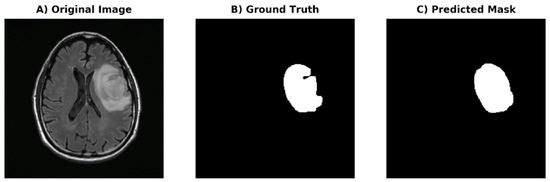
U-Net Based Segmentation and Characterization of Gliomas
by
Shingo Kihira, Xueyan Mei, Keon Mahmoudi, Zelong Liu, Siddhant Dogra, Puneet Belani, Nadejda Tsankova, Adilia Hormigo, Zahi A. Fayad, Amish Doshi and Kambiz Nael
Cited by 10 | Viewed by 2079
Abstract
(1) Background: Gliomas are the most common primary brain neoplasms accounting for roughly 40–50% of all malignant primary central nervous system tumors. We aim to develop a deep learning-based framework for automated segmentation and prediction of biomarkers and prognosis in patients with gliomas.
[...] Read more.
(1) Background: Gliomas are the most common primary brain neoplasms accounting for roughly 40–50% of all malignant primary central nervous system tumors. We aim to develop a deep learning-based framework for automated segmentation and prediction of biomarkers and prognosis in patients with gliomas. (2) Methods: In this retrospective two center study, patients were included if they (1) had a diagnosis of glioma with known surgical histopathology and (2) had preoperative MRI with FLAIR sequence. The entire tumor volume including FLAIR hyperintense infiltrative component and necrotic and cystic components was segmented. Deep learning-based U-Net framework was developed based on symmetric architecture from the 512 × 512 segmented maps from FLAIR as the ground truth mask. (3) Results: The final cohort consisted of 208 patients with mean ± standard deviation of age (years) of 56 ± 15 with M/F of 130/78. DSC of the generated mask was 0.93. Prediction for IDH-1 and MGMT status had a performance of AUC 0.88 and 0.62, respectively. Survival prediction of <18 months demonstrated AUC of 0.75. (4) Conclusions: Our deep learning-based framework can detect and segment gliomas with excellent performance for the prediction of IDH-1 biomarker status and survival.
Full article
►▼
Show Figures
Open AccessArticle
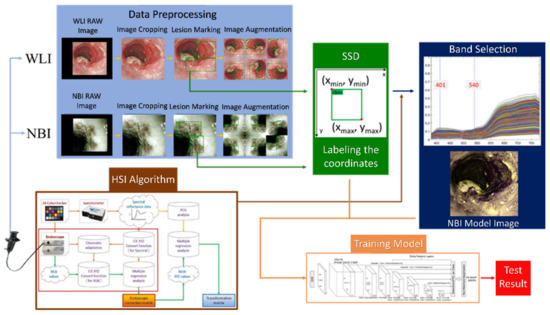
Intelligent Identification of Early Esophageal Cancer by Band-Selective Hyperspectral Imaging
by
Tsung-Jung Tsai, Arvind Mukundan, Yu-Sheng Chi, Yu-Ming Tsao, Yao-Kuang Wang, Tsung-Hsien Chen, I-Chen Wu, Chien-Wei Huang and Hsiang-Chen Wang
Cited by 28 | Viewed by 2497
Abstract
In this study, the combination of hyperspectral imaging (HSI) technology and band selection was coupled with color reproduction. The white-light images (WLIs) were simulated as narrow-band endoscopic images (NBIs). As a result, the blood vessel features in the endoscopic image became more noticeable,
[...] Read more.
In this study, the combination of hyperspectral imaging (HSI) technology and band selection was coupled with color reproduction. The white-light images (WLIs) were simulated as narrow-band endoscopic images (NBIs). As a result, the blood vessel features in the endoscopic image became more noticeable, and the prediction performance was improved. In addition, a single-shot multi-box detector model for predicting the stage and location of esophageal cancer was developed to evaluate the results. A total of 1780 esophageal cancer images, including 845 WLIs and 935 NBIs, were used in this study. The images were divided into three stages based on the pathological features of esophageal cancer: normal, dysplasia, and squamous cell carcinoma. The results showed that the mean average precision (mAP) reached 80% in WLIs, 85% in NBIs, and 84% in HSI images. This study′s results showed that HSI has more spectral features than white-light imagery, and it improves accuracy by about 5% and matches the results of NBI predictions.
Full article
►▼
Show Figures
Open AccessArticle
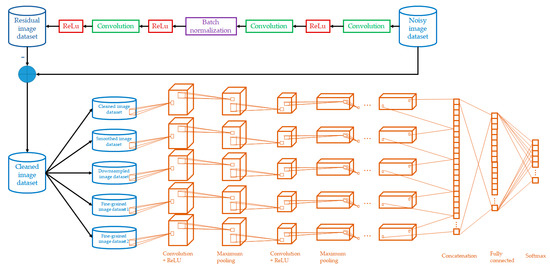
Transfer Learning-Based Multi-Scale Denoising Convolutional Neural Network for Prostate Cancer Detection
by
Kwok Tai Chui, Brij B. Gupta, Hao Ran Chi, Varsha Arya, Wadee Alhalabi, Miguel Torres Ruiz and Chien-Wen Shen
Cited by 17 | Viewed by 1964
Abstract
Background: Prostate cancer is the 4th most common type of cancer. To reduce the workload of medical personnel in the medical diagnosis of prostate cancer and increase the diagnostic accuracy in noisy images, a deep learning model is desired for prostate cancer detection.
[...] Read more.
Background: Prostate cancer is the 4th most common type of cancer. To reduce the workload of medical personnel in the medical diagnosis of prostate cancer and increase the diagnostic accuracy in noisy images, a deep learning model is desired for prostate cancer detection. Methods: A multi-scale denoising convolutional neural network (MSDCNN) model was designed for prostate cancer detection (PCD) that is capable of noise suppression in images. The model was further optimized by transfer learning, which contributes domain knowledge from the same domain (prostate cancer data) but heterogeneous datasets. Particularly, Gaussian noise was introduced in the source datasets before knowledge transfer to the target dataset. Results: Four benchmark datasets were chosen as representative prostate cancer datasets. Ablation study and performance comparison between the proposed work and existing works were performed. Our model improved the accuracy by more than 10% compared with the existing works. Ablation studies also showed average improvements in accuracy using denoising, multi-scale scheme, and transfer learning, by 2.80%, 3.30%, and 3.13%, respectively. Conclusions: The performance evaluation and comparison of the proposed model confirm the importance and benefits of image noise suppression and transfer of knowledge from heterogeneous datasets of the same domain.
Full article
►▼
Show Figures
Open AccessArticle
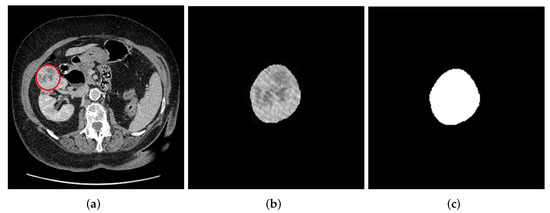
Comparative Analysis for the Distinction of Chromophobe Renal Cell Carcinoma from Renal Oncocytoma in Computed Tomography Imaging Using Machine Learning Radiomics Analysis
by
Abeer J. Alhussaini, J. Douglas Steele and Ghulam Nabi
Cited by 7 | Viewed by 4097
Abstract
Background: ChRCC and RO are two types of rarely occurring renal tumors that are difficult to distinguish from one another based on morphological features alone. They differ in prognosis, with ChRCC capable of progressing and metastasizing, but RO is benign. This means discrimination
[...] Read more.
Background: ChRCC and RO are two types of rarely occurring renal tumors that are difficult to distinguish from one another based on morphological features alone. They differ in prognosis, with ChRCC capable of progressing and metastasizing, but RO is benign. This means discrimination of the two tumors is of crucial importance.
Objectives: The purpose of this research was to develop and comprehensively evaluate predictive models that can discriminate between ChRCC and RO tumors using Computed Tomography (CT) scans and ML-Radiomics texture analysis methods.
Methods: Data were obtained from 78 pathologically confirmed renal masses, scanned at two institutions. Data from the two institutions were combined to form a third set resulting in three data cohorts, i.e., cohort 1, 2 and combined. Contrast-enhanced scans were used and the axial cross-sectional slices of each tumor were extracted from the 3D data using a semi-automatic segmentation technique for both 2D and 3D scans. Radiomics features were extracted before and after applying filters and the dimensions of the radiomic features reduced using the least absolute shrinkage and selection operator (LASSO) method. Synthetic minority oversampling technique (SMOTE) was applied to avoid class imbalance. Five ML algorithms were used to train models for predictive classification and evaluated using 5-fold cross-validation.
Results: The number of selected features with good model performance was 20, 40 and 6 for cohorts 1, 2 and combined, respectively. The best model performance in cohorts 1, 2 and combined had an excellent Area Under the Curve (AUC) of 1.00 ± 0.000, 1.00 ± 0.000 and 0.87 ± 0.073, respectively. Conclusions: ML-based radiomics signatures are potentially useful for distinguishing ChRCC and RO tumors, with a reliable level of performance for both 2D and 3D scanning.
Full article
►▼
Show Figures
Open AccessArticle
Early Prediction of Planning Adaptation Requirement Indication Due to Volumetric Alterations in Head and Neck Cancer Radiotherapy: A Machine Learning Approach
by
Vasiliki Iliadou, Ioannis Kakkos, Pantelis Karaiskos, Vassilis Kouloulias, Kalliopi Platoni, Anna Zygogianni and George K. Matsopoulos
Cited by 6 | Viewed by 1985
Abstract
Background: During RT cycles, the tumor response pattern could affect tumor coverage and may lead to organs at risk of overdose. As such, early prediction of significant volumetric changes could therefore reduce potential radiation-related adverse effects. Nevertheless, effective machine learning approaches based on
[...] Read more.
Background: During RT cycles, the tumor response pattern could affect tumor coverage and may lead to organs at risk of overdose. As such, early prediction of significant volumetric changes could therefore reduce potential radiation-related adverse effects. Nevertheless, effective machine learning approaches based on the radiomic features of the clinically used CBCT images to determine the tumor volume variations due to RT not having been implemented so far. Methods: CBCT images from 40 HN cancer patients were collected weekly during RT treatment. From the obtained images, the Clinical Target Volume (CTV) and Parotid Glands (PG) regions of interest were utilized to calculate 104 delta-radiomics features. These features were fed on a feature selection and classification procedure for the early prediction of significant volumetric alterations. Results: The proposed framework was able to achieve 0.90 classification performance accuracy while detecting a small subset of discriminative characteristics from the 1st week of RT. The selected features were further analyzed regarding their effects on temporal changes in anatomy and tumor response modeling. Conclusion: The use of machine learning algorithms offers promising perspectives for fast and reliable early prediction of large volumetric deviations as a result of RT treatment, exploiting hidden patterns in the overall anatomical characteristics.
Full article
►▼
Show Figures
Open AccessArticle
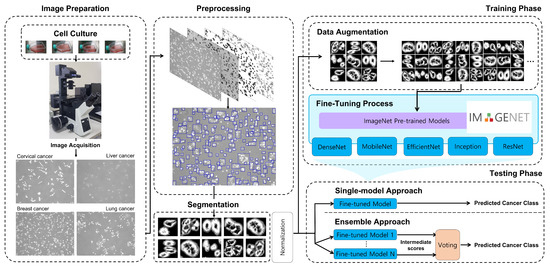
Automatic Cancer Cell Taxonomy Using an Ensemble of Deep Neural Networks
by
Se-woon Choe, Ha-Yeong Yoon, Jae-Yeop Jeong, Jinhyung Park and Jin-Woo Jeong
Cited by 4 | Viewed by 2281
Abstract
Microscopic image-based analysis has been intensively performed for pathological studies and diagnosis of diseases. However, mis-authentication of cell lines due to misjudgments by pathologists has been recognized as a serious problem. To address this problem, we propose a deep-learning-based approach for the automatic
[...] Read more.
Microscopic image-based analysis has been intensively performed for pathological studies and diagnosis of diseases. However, mis-authentication of cell lines due to misjudgments by pathologists has been recognized as a serious problem. To address this problem, we propose a deep-learning-based approach for the automatic taxonomy of cancer cell types. A total of 889 bright-field microscopic images of four cancer cell lines were acquired using a benchtop microscope. Individual cells were further segmented and augmented to increase the image dataset. Afterward, deep transfer learning was adopted to accelerate the classification of cancer types. Experiments revealed that the deep-learning-based methods outperformed traditional machine-learning-based methods. Moreover, the Wilcoxon signed-rank test showed that deep ensemble approaches outperformed individual deep-learning-based models (
p < 0.001) and were in effect to achieve the classification accuracy up to 97.735%. Additional investigation with the Wilcoxon signed-rank test was conducted to consider various network design choices, such as the type of optimizer, type of learning rate scheduler, degree of fine-tuning, and use of data augmentation. Finally, it was found that the using data augmentation and updating all the weights of a network during fine-tuning improve the overall performance of individual convolutional neural network models.
Full article
►▼
Show Figures
Open AccessArticle
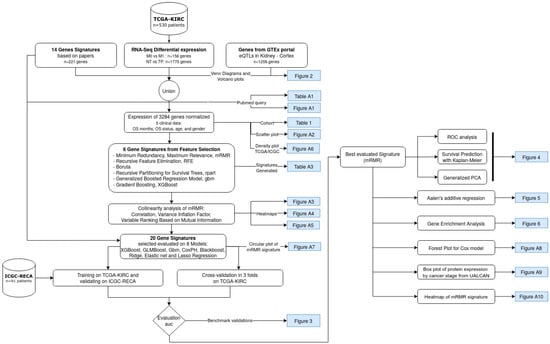
A Novel Machine Learning 13-Gene Signature: Improving Risk Analysis and Survival Prediction for Clear Cell Renal Cell Carcinoma Patients
by
Patrick Terrematte, Dhiego Souto Andrade, Josivan Justino, Beatriz Stransky, Daniel Sabino A. de Araújo and Adrião D. Dória Neto
Cited by 12 | Viewed by 3854
Abstract
Patients with clear cell renal cell carcinoma (ccRCC) have poor survival outcomes, especially if it has metastasized. It is of paramount importance to identify biomarkers in genomic data that could help predict the aggressiveness of ccRCC and its resistance to drugs. Thus, we
[...] Read more.
Patients with clear cell renal cell carcinoma (ccRCC) have poor survival outcomes, especially if it has metastasized. It is of paramount importance to identify biomarkers in genomic data that could help predict the aggressiveness of ccRCC and its resistance to drugs. Thus, we conducted a study with the aims of evaluating gene signatures and proposing a novel one with higher predictive power and generalization in comparison to the former signatures. Using ccRCC cohorts of the Cancer Genome Atlas (TCGA-KIRC) and International Cancer Genome Consortium (ICGC-RECA), we evaluated linear survival models of Cox regression with 14 signatures and six methods of feature selection, and performed functional analysis and differential gene expression approaches. In this study, we established a 13-gene signature (AR, AL353637.1, DPP6, FOXJ1, GNB3, HHLA2, IL4, LIMCH1, LINC01732, OTX1, SAA1, SEMA3G, ZIC2) whose expression levels are able to predict distinct outcomes of patients with ccRCC. Moreover, we performed a comparison between our signature and others from the literature. The best-performing gene signature was achieved using the ensemble method Min-Redundancy and Max-Relevance (mRMR). This signature comprises unique features in comparison to the others, such as generalization through different cohorts and being functionally enriched in significant pathways: Urothelial Carcinoma, Chronic Kidney disease, and Transitional cell carcinoma, Nephrolithiasis. From the 13 genes in our signature, eight are known to be correlated with ccRCC patient survival and four are immune-related. Our model showed a performance of 0.82 using the Receiver Operator Characteristic (ROC) Area Under Curve (AUC) metric and it generalized well between the cohorts. Our findings revealed two clusters of genes with high expression (SAA1, OTX1, ZIC2, LINC01732, GNB3 and IL4) and low expression (AL353637.1, AR, HHLA2, LIMCH1, SEMA3G, DPP6, and FOXJ1) which are both correlated with poor prognosis. This signature can potentially be used in clinical practice to support patient treatment care and follow-up.
Full article
►▼
Show Figures
Open AccessArticle
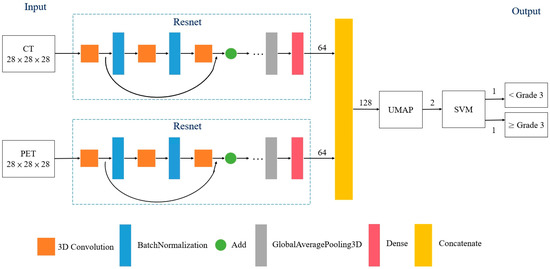
Prediction of Neoadjuvant Chemoradiotherapy Response in Rectal Cancer with Metric Learning Using Pretreatment 18F-Fluorodeoxyglucose Positron Emission Tomography
by
Kuo-Chen Wu, Shang-Wen Chen, Te-Chun Hsieh, Kuo-Yang Yen, Kin-Man Law, Yu-Chieh Kuo, Ruey-Feng Chang and Chia-Hung Kao
Cited by 2 | Viewed by 2068
Abstract
Objectives: Neoadjuvant chemoradiotherapy (NCRT) followed by surgery is the mainstay of treatment for patients with locally advanced rectal cancer. Based on baseline 18F-fluorodeoxyglucose ([18F]-FDG)-positron emission tomography (PET)/computed tomography (CT), a new artificial intelligence model using metric learning (ML) was introduced to predict responses
[...] Read more.
Objectives: Neoadjuvant chemoradiotherapy (NCRT) followed by surgery is the mainstay of treatment for patients with locally advanced rectal cancer. Based on baseline 18F-fluorodeoxyglucose ([18F]-FDG)-positron emission tomography (PET)/computed tomography (CT), a new artificial intelligence model using metric learning (ML) was introduced to predict responses to NCRT. Patients and Methods: This study used the data of 236 patients with newly diagnosed rectal cancer; the data of 202 and 34 patients were for training and validation, respectively. All patients received pretreatment [18F]FDG-PET/CT, NCRT, and surgery. The treatment response was scored by Dworak tumor regression grade (TRG); TRG3 and TRG4 indicated favorable responses. The model employed ML combined with the Uniform Manifold Approximation and Projection for dimensionality reduction. A receiver operating characteristic (ROC) curve analysis was performed to assess the model’s predictive performance. Results: In the training cohort, 115 patients (57%) achieved TRG3 or TRG4 responses. The area under the ROC curve was 0.96 for the prediction of a favorable response. The sensitivity, specificity, and accuracy were 98.3%, 96.5%, and 97.5%, respectively. The sensitivity, specificity, and accuracy for the validation cohort were 95.0%, 100%, and 98.8%, respectively. Conclusions: The new ML model presented herein was used to determined that baseline 18F[FDG]-PET/CT images could predict a favorable response to NCRT in patients with rectal cancer. External validation is required to verify the model’s predictive value.
Full article
►▼
Show Figures
Open AccessArticle
Machine Learning for Prediction of Immunotherapy Efficacy in Non-Small Cell Lung Cancer from Simple Clinical and Biological Data
by
Sébastien Benzekry, Mathieu Grangeon, Mélanie Karlsen, Maria Alexa, Isabella Bicalho-Frazeto, Solène Chaleat, Pascale Tomasini, Dominique Barbolosi, Fabrice Barlesi and Laurent Greillier
Cited by 16 | Viewed by 4095
Abstract
Background: Immune checkpoint inhibitors (ICIs) are now a therapeutic standard in advanced non-small cell lung cancer (NSCLC), but strong predictive markers for ICIs efficacy are still lacking. We evaluated machine learning models built on simple clinical and biological data to individually predict response
[...] Read more.
Background: Immune checkpoint inhibitors (ICIs) are now a therapeutic standard in advanced non-small cell lung cancer (NSCLC), but strong predictive markers for ICIs efficacy are still lacking. We evaluated machine learning models built on simple clinical and biological data to individually predict response to ICIs. Methods: Patients with metastatic NSCLC who received ICI in second line or later were included. We collected clinical and hematological data and studied the association of this data with disease control rate (DCR), progression free survival (PFS) and overall survival (OS). Multiple machine learning (ML) algorithms were assessed for their ability to predict response. Results: Overall, 298 patients were enrolled. The overall response rate and DCR were 15.3% and 53%, respectively. Median PFS and OS were 3.3 and 11.4 months, respectively. In multivariable analysis, DCR was significantly associated with performance status (PS) and hemoglobin level (OR 0.58,
p < 0.0001; OR 1.8,
p < 0.001). These variables were also associated with PFS and OS and ranked top in random forest-based feature importance. Neutrophil-to-lymphocyte ratio was also associated with DCR, PFS and OS. The best ML algorithm was a random forest. It could predict DCR with satisfactory efficacy based on these three variables. Ten-fold cross-validated performances were: accuracy 0.68 ± 0.04, sensitivity 0.58 ± 0.08; specificity 0.78 ± 0.06; positive predictive value 0.70 ± 0.08; negative predictive value 0.68 ± 0.06; AUC 0.74 ± 0.03. Conclusion: Combination of simple clinical and biological data could accurately predict disease control rate at the individual level.
Full article
►▼
Show Figures
Open AccessArticle
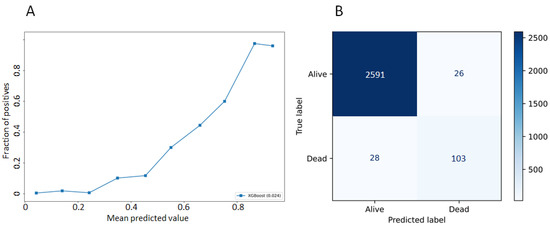
Development and Validation of an Interpretable Artificial Intelligence Model to Predict 10-Year Prostate Cancer Mortality
by
Jean-Emmanuel Bibault, Steven Hancock, Mark K. Buyyounouski, Hilary Bagshaw, John T. Leppert, Joseph C. Liao and Lei Xing
Cited by 8 | Viewed by 3181
Abstract
Prostate cancer treatment strategies are guided by risk-stratification. This stratification can be difficult in some patients with known comorbidities. New models are needed to guide strategies and determine which patients are at risk of prostate cancer mortality. This article presents a gradient-boosting model
[...] Read more.
Prostate cancer treatment strategies are guided by risk-stratification. This stratification can be difficult in some patients with known comorbidities. New models are needed to guide strategies and determine which patients are at risk of prostate cancer mortality. This article presents a gradient-boosting model to predict the risk of prostate cancer mortality within 10 years after a cancer diagnosis, and to provide an interpretable prediction. This work uses prospective data from the PLCO Cancer Screening and selected patients who were diagnosed with prostate cancer. During follow-up, 8776 patients were diagnosed with prostate cancer. The dataset was randomly split into a training (
n = 7021) and testing (
n = 1755) dataset. Accuracy was 0.98 (±0.01), and the area under the receiver operating characteristic was 0.80 (±0.04). This model can be used to support informed decision-making in prostate cancer treatment. AI interpretability provides a novel understanding of the predictions to the users.
Full article
►▼
Show Figures