Journal Description
Biophysica
Biophysica
is an international, peer-reviewed, open access journal on applying the methods of physics, chemistry, and math to study biological systems, published quarterly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, EBSCO, and other databases.
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 16.7 days after submission; acceptance to publication is undertaken in 3.7 days (median values for papers published in this journal in the second half of 2023).
- Recognition of reviewers: APC discount vouchers, optional signed peer review and reviewer names are published annually in the journal.
- Biophysica is a companion journal of IJMS.
Latest Articles
Biophysical Breakthroughs Projected for the Phage Therapy of Bacterial Disease
Biophysica 2024, 4(2), 195-206; https://doi.org/10.3390/biophysica4020014 - 12 Apr 2024
Abstract
►
Show Figures
Past anti-bacterial use of bacteriophages (phage therapy) is already well reviewed as a potential therapeutic response to the emergence of multidrug-resistant, pathogenic bacteria. Phage therapy has been limited by the following. (1) The success rate is too low for routine use and Food
[...] Read more.
Past anti-bacterial use of bacteriophages (phage therapy) is already well reviewed as a potential therapeutic response to the emergence of multidrug-resistant, pathogenic bacteria. Phage therapy has been limited by the following. (1) The success rate is too low for routine use and Food and Drug Administration (FDA) approval. (2) Current strategies of routine phage characterization do not sufficiently improve the success rate of phage therapy. (3) The stability of many phages at ambient temperature is not high enough to routinely store and transport phages at ambient temperature. In the present communication, we present new and previous data that we interpret as introductory to biophysically and efficiently transforming phage therapy to the needed level of effectiveness. Included are (1) procedure and preliminary data for the use of native gel electrophoresis (a low-cost procedure) for projecting the therapy effectiveness of a newly isolated phage, (2) data that suggest a way to achieve stabilizing of dried, ambient-temperature phages via polymer embedding, and (3) data that suggest means to increase the blood persistence, and therefore the therapy effectiveness, of what would otherwise be a relatively low-persistence phage.
Full article
Open AccessArticle
Deciphering the Molecular Interaction Process of Gallium Maltolate on SARS-CoV-2 Main and Papain-Like Proteases: A Theoretical Study
by
Kevin Taype-Huanca, Manuel I. Osorio, Diego Inostroza, Luis Leyva-Parra, Lina Ruíz, Ana Valderrama-Negrón, Jesús Alvarado-Huayhuaz, Osvaldo Yañez and William Tiznado
Biophysica 2024, 4(2), 182-194; https://doi.org/10.3390/biophysica4020013 - 10 Apr 2024
Abstract
This study explored the inhibitory potential of gallium maltolate against severe acute respiratory syndrome coronavirus 2 and main and papain-like proteases. Computational methods, including density functional theory and molecular docking, were used to assess gallium maltolate reactivity and binding interactions. Density functional theory
[...] Read more.
This study explored the inhibitory potential of gallium maltolate against severe acute respiratory syndrome coronavirus 2 and main and papain-like proteases. Computational methods, including density functional theory and molecular docking, were used to assess gallium maltolate reactivity and binding interactions. Density functional theory calculations revealed gallium maltolate’s high electron-capturing capacity, particularly around the gallium metal atom, which may contribute to their activity. Molecular docking demonstrated that gallium maltolate can form strong hydrogen bonds with key amino acid residues like glutamate-166 and cysteine-145, tightly binding to main and papain-like proteases. The binding energy and interactions of gallium maltolate were comparable to known SARS-CoV-2 inhibitors like N-[(5-methyl-1,2-oxazol-3-yl)carbonyl]-L-alanyl-L-valyl-N-{(2S,3E)-5-(benzyloxy)-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pent-3-en-2-yl}-L-leucinamide, indicating its potential as an antiviral agent. However, further experimental validation is required to confirm its effectiveness in inhibiting SARS-CoV-2 replication and treating COVID-19.
Full article
(This article belongs to the Special Issue The Structure and Function of Proteins, Lipids, and Nucleic Acids)
►▼
Show Figures

Figure 1
Open AccessArticle
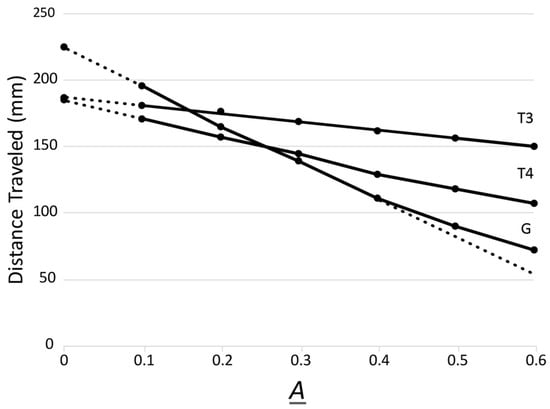
Search for Entanglement between Spatially Separated Living Systems: Experiment Design, Results, and Lessons Learned
by
Chris Fields, Lorenzo Cohen, Andrew Cusimano, Sharmistha Chakraborty, Phuong Nguyen, Defeng Deng, Shafaqmuhammad Iqbal, Monica Nelson, Daoyan Wei, Arnaud Delorme and Peiying Yang
Biophysica 2024, 4(2), 168-181; https://doi.org/10.3390/biophysica4020012 - 30 Mar 2024
Abstract
►▼
Show Figures
Statistically significant violations of the Clauser–Horne–Shimony–Holt (CHSH) inequality are the “gold standard” test for quantum entanglement between spatially separated systems. Here, we report an experimental design that implements a CHSH test between bioelectric state variables for a human subject and bioelectric and/or biochemical
[...] Read more.
Statistically significant violations of the Clauser–Horne–Shimony–Holt (CHSH) inequality are the “gold standard” test for quantum entanglement between spatially separated systems. Here, we report an experimental design that implements a CHSH test between bioelectric state variables for a human subject and bioelectric and/or biochemical state variables for cultured human cells in vitro. While we were unable to obtain evidence for entanglement with this design, observing only classical correlation, we report lessons learned and suggest possible avenues for future studies.
Full article
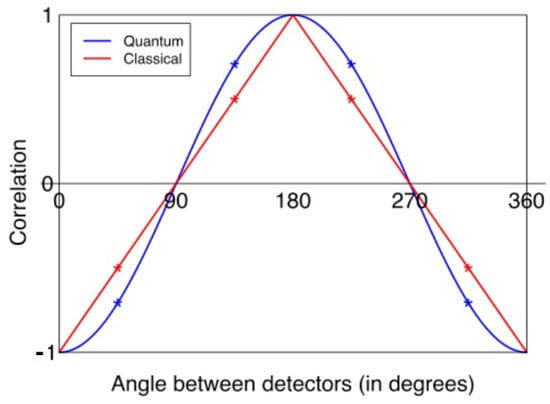
Figure 1
Open AccessPerspective
Never Fold to Fold Continuously: A Conundrum in Ubiquitin–Proteasome System (UPS)-Mediated Protein Quality Control (PQC)
by
Stefano Magnati and Enrico Bracco
Biophysica 2024, 4(2), 158-167; https://doi.org/10.3390/biophysica4020011 - 30 Mar 2024
Abstract
In the last few decades, the traditional paradigm of teleonomy, in which the amino acid sequence of a protein is tightly associated with its structure and, in turn, with its function, has been partially undermined. The idea of a protein as a two-state
[...] Read more.
In the last few decades, the traditional paradigm of teleonomy, in which the amino acid sequence of a protein is tightly associated with its structure and, in turn, with its function, has been partially undermined. The idea of a protein as a two-state object has been superseded by that of understanding it as a multistate object. Indeed, some proteins, or portions of a protein, display intrinsically disordered regions (IDRs), which means that they lack stable secondary or tertiary structures. While we are aware that IDRs are present in almost half of the total human proteins, we are still quite far away from understanding their contextual-specific functions and figuring out how they mechanistically work. In the present perspective article, we will attempt to summarize the role/s of IDRs in ubiquitin–proteasome system (UPS)-mediated protein quality control (PQC) at different levels, ranging from ubiquitination to protein degradation through the proteasome machinery up to their role in decoding the complex ubiquitin code. Ultimately, we will critically discuss the future challenges we are facing to gain insights into the role of IDRs in regulating UPS-mediated PQC.
Full article
(This article belongs to the Special Issue Protein Disorder)
►▼
Show Figures

Figure 1
Open AccessReview
Axial Tomography in Live Cell Microscopy
by
Herbert Schneckenburger and Christoph Cremer
Biophysica 2024, 4(2), 142-157; https://doi.org/10.3390/biophysica4020010 - 29 Mar 2024
Abstract
For many biomedical applications, laser-assisted methods are essential to enhance the three-dimensional (3D) resolution of a light microscope. In this report, we review possibilities to improve the 3D imaging potential by axial tomography. This method allows us to rotate the object in a
[...] Read more.
For many biomedical applications, laser-assisted methods are essential to enhance the three-dimensional (3D) resolution of a light microscope. In this report, we review possibilities to improve the 3D imaging potential by axial tomography. This method allows us to rotate the object in a microscope into the best perspective required for imaging. Furthermore, images recorded under variable angles can be combined to one image with isotropic resolution. After a brief review of the technical state of the art, we show some biomedical applications, and discuss future perspectives for Deep View Microscopy and Molecular Imaging.
Full article
(This article belongs to the Special Issue Biomedical Optics 2.0)
►▼
Show Figures

Figure 1
Open AccessReview
Assessing the Impact of Agents with Antiviral Activities on Transmembrane Ionic Currents: Exploring Possible Unintended Actions
by
Geng-Bai Lin, Chia-Lung Shih, Rasa Liutkevičienė, Vita Rovite, Edmund Cheung So, Chao-Liang Wu and Sheng-Nan Wu
Biophysica 2024, 4(2), 128-141; https://doi.org/10.3390/biophysica4020009 - 27 Mar 2024
Abstract
►▼
Show Figures
As the need for effective antiviral treatment intensifies, such as with the coronavirus disease 19 (COVID-19) infection, it is crucial to understand that while the mechanisms of action of these drugs or compounds seem apparent, they might also interact with unexplored targets, such
[...] Read more.
As the need for effective antiviral treatment intensifies, such as with the coronavirus disease 19 (COVID-19) infection, it is crucial to understand that while the mechanisms of action of these drugs or compounds seem apparent, they might also interact with unexplored targets, such as cell membrane ion channels in diverse cell types. In this review paper, we demonstrate that many different drugs or compounds, in addition to their known interference with viral infections, may also directly influence various types of ionic currents on the surface membrane of the host cell. These agents include artemisinin, cannabidiol, memantine, mitoxantrone, molnupiravir, remdesivir, SM-102, and sorafenib. If achievable at low concentrations, these regulatory effects on ion channels are highly likely to synergize with the identified initial mechanisms of viral replication interference. Additionally, the immediate regulatory impact of these agents on the ion-channel function may potentially result in unintended adverse effects, including changes in cardiac electrical activity and the prolongation of the QTc interval. Therefore, it is essential for patients receiving these related agents to exercise additional caution to prevent unnecessary complications.
Full article
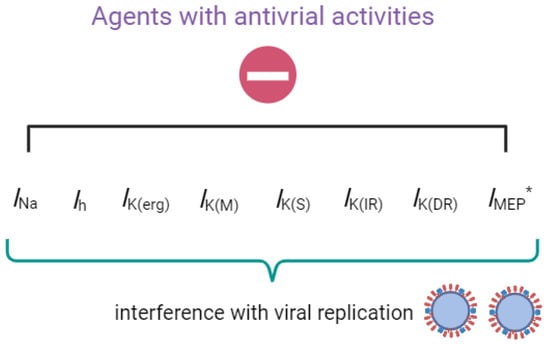
Figure 1
Open AccessArticle
Constant-pH Simulations of a Coarse-Grained Model of Polyfunctional Weak Charged Biopolymers
by
David Naranjo, Pablo M. Blanco, Josep L. Garcés, Sergio Madurga and Francesc Mas
Biophysica 2024, 4(1), 107-127; https://doi.org/10.3390/biophysica4010008 - 28 Feb 2024
Abstract
A coarse-grained model of linear polyfunctional weak charged biopolymers was implemented, formed of different proportions of acid-base groups resembling the composition of humic substances. These substances are mainly present in dissolved organic matter in natural water. The influence of electrostatic interactions computing methods,
[...] Read more.
A coarse-grained model of linear polyfunctional weak charged biopolymers was implemented, formed of different proportions of acid-base groups resembling the composition of humic substances. These substances are mainly present in dissolved organic matter in natural water. The influence of electrostatic interactions computing methods, factors concerning the structure of the chain, different functional groups, and the ionic strength on polyelectrolytes were studied. Langevin dynamics with constant pH simulations were performed using the ESPResSO package and the Python-based Molecule Builder for ESPResSo (pyMBE) library. The coverage was fitted to a polyfunctional Frumkin isotherm, with a mean-field interaction between charged beads. The composition of the chain affects the charge while ionic strength affects both the charge and the radius of gyration. Additionally, the parameters intrinsic to the polyelectrolyte model were well reproduced by fitting the polyfunctional Frumkin isotherm. In contrast, the non-intrinsic parameters depended on the ionic strength. The method developed and applied to a polyfunctional polypeptide model, that resembles a humic acid, will be very useful for characterizing biopolymers with several acid-base functional groups, where their structure, the composition of the different functional groups, and the determination of the main intrinsic proton binding constants and their proportion are not exactly known.
Full article
(This article belongs to the Special Issue Molecular Structure and Simulation in Biological System 2.0)
►▼
Show Figures
Figure 1
Open AccessArticle
The Signature of Fluctuations of the Hydrogen Bond Network Formed by Water Molecules in the Interfacial Layer of Anionic Lipids
by
Ana-Marija Pavlek, Barbara Pem and Danijela Bakarić
Biophysica 2024, 4(1), 92-106; https://doi.org/10.3390/biophysica4010007 - 21 Feb 2024
Abstract
As the water molecules found at the interface of lipid bilayers exhibit distinct structural and reorientation dynamics compared to water molecules found in bulk, the fluctuations in their hydrogen bond (HB) network are expected to be different from those generated by the bulk
[...] Read more.
As the water molecules found at the interface of lipid bilayers exhibit distinct structural and reorientation dynamics compared to water molecules found in bulk, the fluctuations in their hydrogen bond (HB) network are expected to be different from those generated by the bulk water molecules. The research presented here aims to gain an insight into temperature-dependent fluctuations of a HB network of water molecules found in an interfacial layer of multilamellar liposomes (MLVs) composed of anionic 1,2-dimyristoyl-sn-glycero-3-phospho-L-serine (DMPS) lipids. Besides suspending DMPS lipids in phosphate buffer saline (PBS) of different pH values (6.0, 7.4, and 8.0), the changes in HB network fluctuations were altered by the incorporation of a non-polar flavonoid molecule myricetin (MCE) within the hydrocarbon chain region. By performing a multivariate analysis on the water combination band observed in temperature-dependent FTIR spectra, the results of which were further mathematically analyzed, the temperature-dependent fluctuations of interfacial water molecules were captured; the latter were the greatest for DMPS in PBS with a pH value of 7.4 and in general were greater for DMPS multibilayers in the absence of MCE. The presence of MCE made DMPS lipids more separated, allowing deeper penetration of water molecules towards the non-polar region and their restricted motion that resulted in decreased fluctuations. The experimentally observed results were supported by MD simulations of DMPS (+MCE) lipid bilayers.
Full article
(This article belongs to the Collection Feature Papers in Biophysics)
►▼
Show Figures
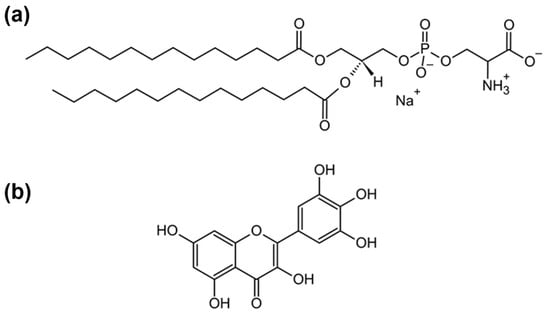
Figure 1
Open AccessCommunication
Direct Interaction of Zirconia Nanoparticles with Human Immune Cells
by
Anna M. Barbasz and Barbara Dyba
Biophysica 2024, 4(1), 83-91; https://doi.org/10.3390/biophysica4010006 - 14 Feb 2024
Abstract
Nanomaterials play a crucial role in various aspects of modern life. Zirconia nanoparticles, extensively employed in medicine for fortifying and stabilizing implants in reconstructive medicine, exhibit unique electrical, thermal, catalytic, sensory, optical, and mechanical properties. While these nanoparticles have shown antibacterial activity, they
[...] Read more.
Nanomaterials play a crucial role in various aspects of modern life. Zirconia nanoparticles, extensively employed in medicine for fortifying and stabilizing implants in reconstructive medicine, exhibit unique electrical, thermal, catalytic, sensory, optical, and mechanical properties. While these nanoparticles have shown antibacterial activity, they also exhibit cytotoxic effects on human cells. Our research focuses on understanding how the cells of the human immune system (both the innate response, namely HL-60 and U-937, and the acquired response, namely HUT-78 and COLO-720L) respond to the presence of zirconium (IV) oxide nanoparticles (ZrO2-NPs). Viability tests indicate that ZrO2-NPs exert the highest cytotoxicity on HL-60 > U-937 > HUT-78 > COLO 720L cell lines. Notably, concentrations exceeding 100 μg mL−1 of ZrO2-NPs result in significant cytotoxicity. These nanoparticles readily penetrate the cell membrane, causing mitochondrial damage, and their cytotoxicity is associated with heightened oxidative stress in cells. The use of ZrO2-NP-based materials may pose a risk to immune system cells, the first responders to foreign entities in the body. Biofunctionalizing the surface of ZrO2-NPs could serve as an effective strategy to mitigate cytotoxicity and introduce new properties for biomedical applications.
Full article
(This article belongs to the Special Issue Functional Application of Nanoparticles in Molecular Biology)
►▼
Show Figures
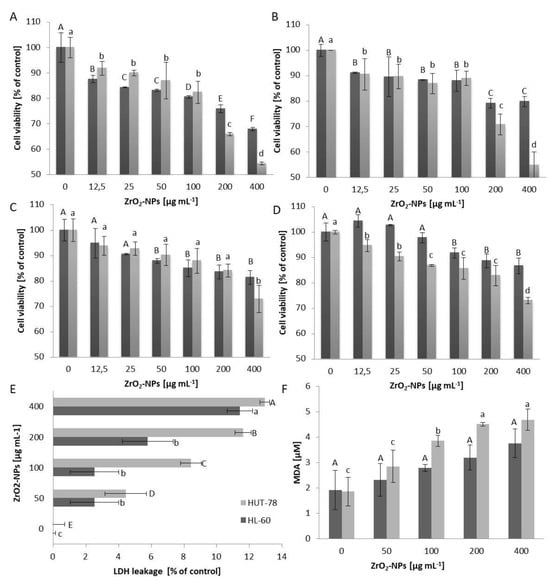
Figure 1
Open AccessCommunication
Neurite Growth and Electrical Activity in PC-12 Cells: Effects of H3 Receptor-Inspired Electromagnetic Fields and Inherent Schumann Frequencies
by
Landon M. Lefebvre, Adam D. Plourde-Kelly, Kevin S. Saroka and Blake T. Dotta
Biophysica 2024, 4(1), 74-82; https://doi.org/10.3390/biophysica4010005 - 07 Feb 2024
Abstract
Cells are continually exposed to a range of electromagnetic fields (EMFs), including those from the Schumann resonance to radio waves. The effects of EMFs on cells are diverse and vary based on the specific EMF type. Recent research suggests potential therapeutic applications of
[...] Read more.
Cells are continually exposed to a range of electromagnetic fields (EMFs), including those from the Schumann resonance to radio waves. The effects of EMFs on cells are diverse and vary based on the specific EMF type. Recent research suggests potential therapeutic applications of EMFs for various diseases. In this study, we explored the impact of a physiologically patterned EMF, inspired by the H3 receptor associated with wakefulness, on PC-12 cells in vitro. Our hypothesis posited that the application of this EMF to differentiated PC-12 cells could enhance firing patterns at specific frequencies. Cell electrophysiology was assessed using a novel device, allowing the computation of spectral power density (SPD) scores for frequencies between 1 Hz and 128 Hz. T-tests comparing SPD at certain frequencies (e.g., 29 Hz, 30 Hz, and 79 Hz) between the H3-EMF and control groups showed a significantly higher SPD in the H3 group (p < 0.050). Moreover, at 7.8 Hz and 71 Hz, a significant correlation was observed between predicted and percentages of cells with neurites (R = 0.542). Key findings indicate the efficacy of the new electrophysiology measure for assessing PC-12 cell activity, a significant increase in cellular activity with the H3-receptor-inspired EMF at specific frequencies, and the influence of 7.8 Hz and 71 Hz frequencies on neurite growth. The overall findings support the idea that the electrical frequency profiles of developing cell systems can serve as an indicator of their progression and eventual cellular outcomes.
Full article
(This article belongs to the Special Issue Biological Effects of Ionizing Radiation)
►▼
Show Figures
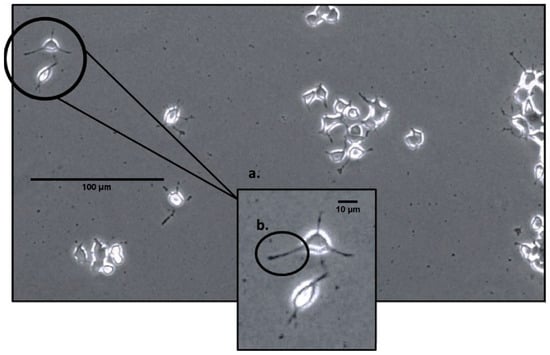
Figure 1
Open AccessReview
Bioprinting of Hydrogel-Based Drug Delivery Systems for Nerve Tissue Regeneration
by
Eliza Marie Steele, Zacheus L. Carr and Emily Dosmar
Biophysica 2024, 4(1), 58-73; https://doi.org/10.3390/biophysica4010004 - 31 Jan 2024
Abstract
Globally, thousands of people are affected by severe nerve injuries or neurodegenerative disorders. These conditions cannot always be cured because nerve tissue either does not regenerate or does so at a slow rate. Therefore, tissue engineering has emerged as a potential treatment approach.
[...] Read more.
Globally, thousands of people are affected by severe nerve injuries or neurodegenerative disorders. These conditions cannot always be cured because nerve tissue either does not regenerate or does so at a slow rate. Therefore, tissue engineering has emerged as a potential treatment approach. This review discusses 3D bioprinting for scaffold manufacturing, highlights the advantages and disadvantages of common bioprinting techniques, describes important considerations for bioinks, biomaterial inks, and scaffolds, and discusses some drug delivery systems. The primary goal of this review is to bring attention to recent advances in nerve tissue engineering and its possible clinical applications in peripheral nerve, spinal cord, and cerebral nerve regeneration. Only studies that use 3D bioprinting or 3D printing to manufacture hydrogel scaffolds and incorporate the sustained release of a drug or growth factor for nerve regeneration are included. This review indicates that 3D printing is a fast and precise scaffold manufacturing technique but requires printing materials with specific properties to be effective in nervous tissue applications. The results indicate that the sustained release of certain drugs and growth factors from scaffolds can significantly improve post-printing cell viability, cell proliferation, adhesion, and differentiation, as well as functional recovery compared with scaffolds alone. However, more in vivo research needs to be conducted before this approach can be used in clinical applications.
Full article
(This article belongs to the Special Issue Molecular Structure and Simulation in Biological System 2.0)
►▼
Show Figures
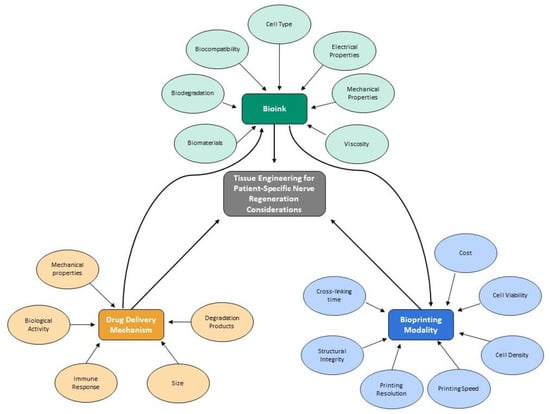
Figure 1
Open AccessArticle
Supercriticality, Glassy Dynamics, and the New Insight into Melting/Freezing Discontinuous Transition in Linseed Oil
by
Aleksandra Drozd-Rzoska, Sylwester J. Rzoska and Joanna Łoś
Biophysica 2024, 4(1), 34-57; https://doi.org/10.3390/biophysica4010003 - 23 Jan 2024
Cited by 1
Abstract
►▼
Show Figures
The long-range supercritical changes of dielectric constant, resembling ones observed in the isotropic liquid phase of liquid crystalline compounds, are evidenced for linseed oil—although in the given case, the phenomenon is associated with the liquid–solid melting/freezing discontinuous phase transitions. This ‘supercriticality’ can be
[...] Read more.
The long-range supercritical changes of dielectric constant, resembling ones observed in the isotropic liquid phase of liquid crystalline compounds, are evidenced for linseed oil—although in the given case, the phenomenon is associated with the liquid–solid melting/freezing discontinuous phase transitions. This ‘supercriticality’ can be an additional factor supporting the unique pro-health properties of linseed oil. Broadband dielectric spectroscopy studies also revealed the ‘glassy’ changes of relaxation times, well portrayed by the ‘activated and critical’ equation recently introduced. In the solid phase, the premelting effect characteristic for the canonic melting/freezing discontinuous transition, i.e., without any pretransitional effect in the liquid phase, has been detected. It is interpreted within the grain model, and its parameterization is possible using the Lipovsky model and the ‘reversed’ Mossotti catastrophe concept. For the premelting effect in the solid state, the singular ‘critical’ temperature correlates with the bulk discontinuous melting and freezing temperatures. Consequently, the report shows that linseed oil, despite its ‘natural and complex’ origins, can be considered a unique model system for two fundamental problems: (i) pretransitional (supercritical) effects in the liquid state associated with a weakly discontinuous phase transition, and (ii) the premelting behavior in the solid side of the discontinuous melting/freezing discontinuous transition.
Full article
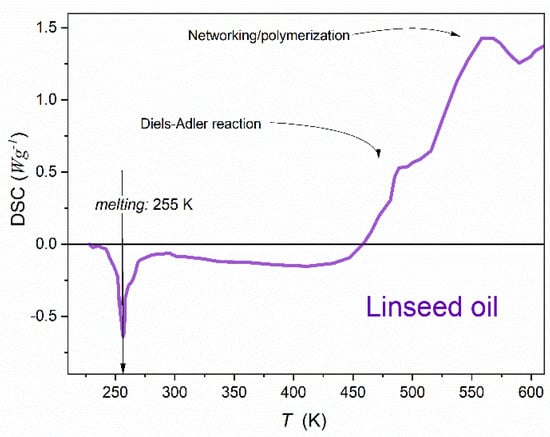
Figure 1
Open AccessArticle
The Effect of UV-Vis Radiation on DNA Systems Containing the Photosensitizers Methylene Blue and Acridine Orange
by
Thais P. Pivetta, Paulo A. Ribeiro and Maria Raposo
Biophysica 2024, 4(1), 22-33; https://doi.org/10.3390/biophysica4010002 - 12 Jan 2024
Abstract
As a vital biomolecule, DNA is known as a target of antineoplastic drugs for cancer therapy. These drugs can show different modes of interaction with DNA, with intercalation and groove binding being the most common types. The intercalation of anticancer drugs with DNA
[...] Read more.
As a vital biomolecule, DNA is known as a target of antineoplastic drugs for cancer therapy. These drugs can show different modes of interaction with DNA, with intercalation and groove binding being the most common types. The intercalation of anticancer drugs with DNA can lead to the disruption of its normal function, influencing cell proliferation. Methylene blue (MB) and acridine orange (AO) are examples of DNA-intercalating agents that have been studied for their application against some types of cancer, mainly for photodynamic therapy. In this work, the impact of light irradiation on these compounds in the absence and presence of DNA was analyzed by means of UV-vis spectroscopy. Bathochromic and hypochromic shifts were observed in the absorbance spectra, revealing the intercalation of the dyes with the DNA base pairs. Dyes with and without DNA present different profiles of photodegradation, whereby the dyes alone were more susceptible to degradation. This can be justified by the intercalation of the dyes on the DNA base pairs allowing the DNA molecule to partially hinder the molecules’ exposition and, therefore, reducing their degradation.
Full article
(This article belongs to the Special Issue Biological Effects of Ionizing Radiation)
►▼
Show Figures
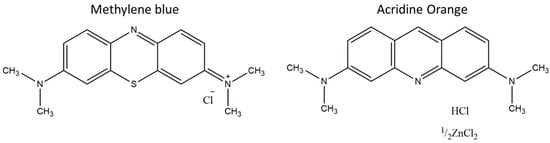
Figure 1
Open AccessReview
The Influence of Magnetic Fields, Including the Planetary Magnetic Field, on Complex Life Forms: How Do Biological Systems Function in This Field and in Electromagnetic Fields?
by
David A. Hart
Biophysica 2024, 4(1), 1-21; https://doi.org/10.3390/biophysica4010001 - 02 Jan 2024
Abstract
►▼
Show Figures
Life on Earth evolved to accommodate the biochemical and biophysical boundary conditions of the planet millions of years ago. The former includes nutrients, water, and the ability to synthesize other needed chemicals. The latter includes the 1 g gravity of the planet, radiation,
[...] Read more.
Life on Earth evolved to accommodate the biochemical and biophysical boundary conditions of the planet millions of years ago. The former includes nutrients, water, and the ability to synthesize other needed chemicals. The latter includes the 1 g gravity of the planet, radiation, and the geomagnetic field (GMF) of the planet. How complex life forms have accommodated the GMF is not known in detail, considering that Homo sapiens evolved a neurological system, a neuromuscular system, and a cardiovascular system that developed electromagnetic fields as part of their functioning. Therefore, all of these could be impacted by magnetic fields. In addition, many proteins and physiologic processes utilize iron ions, which exhibit magnetic properties. Thus, complex organisms, such as humans, generate magnetic fields, contain significant quantities of iron ions, and respond to exogenous static and electromagnetic fields. Given the current body of literature, it remains somewhat unclear if Homo sapiens use exogenous magnetic fields to regulate function and what can happen if the boundary condition of the GMF no longer exerts an effect. Proposed deep space flights to destinations such as Mars will provide some insights, as space flight could not have been anticipated by evolution. The results of such space flight “experiments” will provide new insights into the role of magnetic fields on human functioning. This review will discuss the literature regarding the involvement of magnetic fields in various normal and disturbed processes in humans while on Earth and then further discuss potential outcomes when the GMF is no longer present to impact host systems, as well as the limitations in the current knowledge. The GMF has been present throughout evolution, but many details of its role in human functioning remain to be elucidated, and how humans have adapted to such fields in order to develop and retain function remains to be elucidated. Why this understudied area has not received the attention required to elucidate the critical information remains a conundrum for both health professionals and those embarking on space flight. However, proposed deep space flights to destinations such as Mars may provide the environments to test and assess the potential roles of magnetic fields in human functioning.
Full article
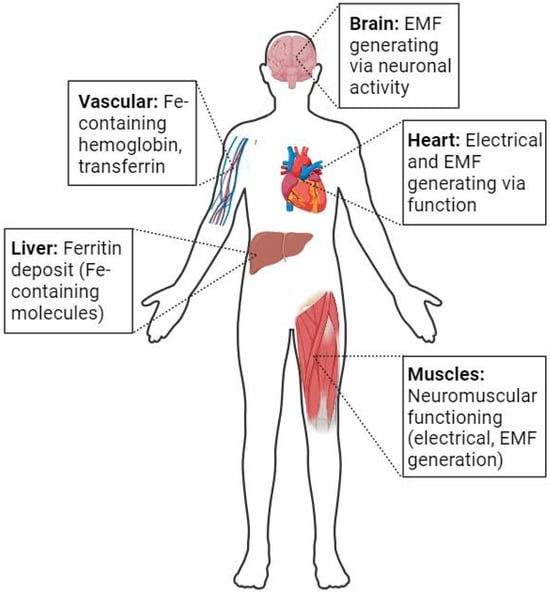
Figure 1
Open AccessOpinion
Does Proprioception Involve Synchronization with Theta Rhythms by a Novel Piezo2 Initiated Ultrafast VGLUT2 Signaling?
by
Balázs Sonkodi
Biophysica 2023, 3(4), 695-710; https://doi.org/10.3390/biophysica3040046 - 18 Dec 2023
Cited by 1
Abstract
This opinion manuscript outlines how the hippocampal theta rhythm could receive two novel peripheral inputs. One of the ways this could be achieved is through Piezo2 channels and atypical hippocampal-like metabotropic glutamate receptors coupled to phospholipase D containing proprioceptive primary afferent terminals. Accordingly,
[...] Read more.
This opinion manuscript outlines how the hippocampal theta rhythm could receive two novel peripheral inputs. One of the ways this could be achieved is through Piezo2 channels and atypical hippocampal-like metabotropic glutamate receptors coupled to phospholipase D containing proprioceptive primary afferent terminals. Accordingly, activated proprioceptive terminal Piezo2 on Type Ia fibers synchronizes to the theta rhythm with the help of hippocampal Piezo2 and medial septal glutamatergic neurons. Second, after baroreceptor Piezo2 is entrained to activated proprioceptive Piezo2, it could turn on the Cav1.3 channels, which pace the heart rhythm and regulate pacemaker cells during cardiac sympathetic activation. This would allow the Cav1.3 channels to synchronize to theta rhythm pacemaker hippocampal parvalbumin-expressing GABAergic neurons. This novel Piezo2-initiated proton–proton frequency coupling through VGLUT2 may provide the ultrafast long-range signaling pathway for the proposed Piezo2 synchronization of the low-frequency glutamatergic cell surface membrane oscillations in order to provide peripheral spatial and speed inputs to the space and speed coding of the hippocampal theta rhythm, supporting locomotion, learning and memory. Moreover, it provides an ultrafast signaling for postural and orthostatic control. Finally, suggestions are made as to how Piezo2 channelopathy could impair this ultrafast communication in many conditions and diseases with not entirely known etiology, leading to impaired proprioception and/or autonomic disbalance.
Full article
(This article belongs to the Special Issue Biological Effects of Ionizing Radiation)
Open AccessPerspective
Dual Nucleosomal Double-Strand Breaks Are the Key Effectors of Curative Radiation Therapy
by
Anders Brahme and Yvonne Lorat
Biophysica 2023, 3(4), 668-694; https://doi.org/10.3390/biophysica3040045 - 14 Dec 2023
Cited by 1
Abstract
Most ionizing radiation produces δ-rays of ≈1 keV that can impart MGy doses to 100 nm3 volumes of DNA. These events can produce severe dual double-strand breaks (DDSBs) on nucleosomes, particularly in dense heterochromatic DNA. This is the most common multiply
[...] Read more.
Most ionizing radiation produces δ-rays of ≈1 keV that can impart MGy doses to 100 nm3 volumes of DNA. These events can produce severe dual double-strand breaks (DDSBs) on nucleosomes, particularly in dense heterochromatic DNA. This is the most common multiply damaged site, and their probabilities determine the biological effectiveness of different types of radiation. We discuss their frequency, effect on cell survival, DNA repair, and imaging by gold nanoparticle tracers and electron microscopy. This new and valuable nanometer resolution information can be used for determining the optimal tumor cure by maximizing therapeutic effects on tumors and minimizing therapeutic effects on normal tissues. The production of DDSBs makes it important to deliver a rather high dose and LET to the tumor (>2.5 Gy/Fr) and at the same time reach approximately 1.8–2.3 Gy of the lowest possible LET per fraction in TP53 intact normal tissues at risk. Therefore, their intrinsic low-dose hyper-sensitivity (LDHS)-related optimal daily fractionation window is utilized. Before full p53 activation of NHEJ and HR repair at ≈½ Gy, the low-dose apoptosis (LDA) and LDHS minimize normal tissue mutation probabilities. Ion therapy should thus ideally produce the lowest possible LET in normal tissues to avoid elevated DDSBs. Helium to boron ions can achieve this with higher-LET Bragg peaks, producing increased tumor DDSB densities. Interestingly, the highest probability of complication-free cure with boron or heavier ions requires a low LET round-up for the last 10–15 GyE, thereby steepening the dose response and further minimizing normal tissue damage. In conclusion, the new high-resolution DSB and DDSB diagnostic methods, and the new more accurate DNA-repair-based radiation biology, have been combined to increase our understanding of what is clinically important in curative radiation therapy. In fact, we must understand that we already passed the region of optimal LET and need to go back one step rather than forward, with oxygen being contemplated. As seen by the high overkill and severely high LET in the distal tumor and the increased LET to normal tissues (reminding of neutrons or neon ions), it is therefore preferable to use lithium–boron ions or combine carbon with an optimal 10–15 GyE photon, electron, or perhaps even a proton round-up, thus allowing optimized, fractionated, curative, almost complication-free treatments with photons, electrons, and light ions, introducing a real paradigm shift in curative radiation therapy with a potential 5 GyE tumor boost, 25% increase in complication-free cure and apoptotic–senescent Bragg Peak molecular light ion radiation therapy.
Full article
(This article belongs to the Special Issue Biological Effects of Ionizing Radiation)
►▼
Show Figures
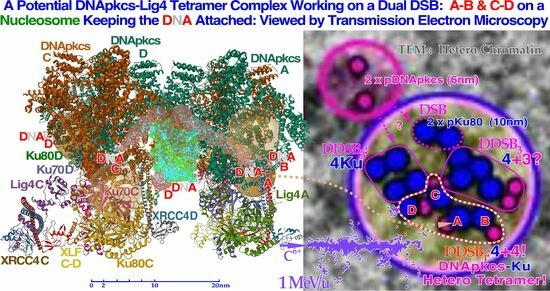
Graphical abstract
Open AccessArticle
Screening and Analysis of Potential Inhibitors of SHMT2
by
Bojin Chen and John Z. H. Zhang
Biophysica 2023, 3(4), 651-667; https://doi.org/10.3390/biophysica3040044 - 03 Dec 2023
Abstract
Serine hydroxymethyltransferase 2 (SHMT2) has garnered significant attention as a critical catalytic regulator of the serine/glycine pathway in the one-carbon metabolism of cancer cells. Despite its potential as an anti-cancer target, only a limited number of inhibitors have been identified so far. In
[...] Read more.
Serine hydroxymethyltransferase 2 (SHMT2) has garnered significant attention as a critical catalytic regulator of the serine/glycine pathway in the one-carbon metabolism of cancer cells. Despite its potential as an anti-cancer target, only a limited number of inhibitors have been identified so far. In this study, we employed seven different scoring functions and skeleton clustering to screen the ChemDiv database for 38 compounds, 20 of which originate from the same skeleton structure. The most significant residues from SHMT2 and chemical groups from the inhibitors were identified using ASGBIE (Alanine Scanning with Generalized Born model and Interaction Entropy), and the binding energy of each residue was quantitatively determined, revealing the essential features of the protein–inhibitor interaction. The two most important contributing residues are TYR105 and TYR106 of the B chain followed by LEU166 and ARG425 of the A chain. The findings will be greatly helpful in developing a thorough comprehension of the binding mechanisms involved in drug–SHMT2 interactions and offer valuable direction for designing more potent inhibitors.
Full article
(This article belongs to the Special Issue Molecular Structure and Simulation in Biological System 2.0)
►▼
Show Figures
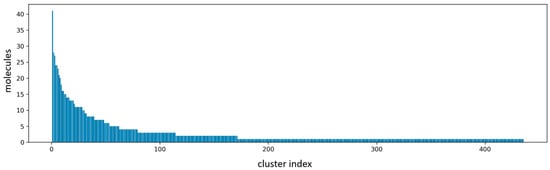
Figure 1
Open AccessArticle
Microgravity as an Anti-Metastatic Agent in an In Vitro Glioma Model
by
Maurizio Sabbatini, Valentina Bonetto, Valeria Magnelli, Candida Lorusso, Francesco Dondero and Maria Angela Masini
Biophysica 2023, 3(4), 636-650; https://doi.org/10.3390/biophysica3040043 - 25 Nov 2023
Abstract
Gravity is a primary physical force that has a profound influence on the stability of the cell cytoskeleton. In our research, we investigated the influence of microgravity on altering the cytoskeletal pathways of glioblastoma cells. The highly infiltrative behavior of glioblastoma is supported
[...] Read more.
Gravity is a primary physical force that has a profound influence on the stability of the cell cytoskeleton. In our research, we investigated the influence of microgravity on altering the cytoskeletal pathways of glioblastoma cells. The highly infiltrative behavior of glioblastoma is supported by cytoskeletal dynamics and surface proteins that allow glioblastoma cells to avoid stable connections with the tissue environment and other cells. Glioblastoma cell line C6 was exposed to a microgravity environment for 24, 48, and 72 h by 3D-RPM, a laboratory instrument recognized to reproduce the effect of microgravity in cell cultures. The immunofluorescence for GFAP, vinculin, and Connexin-43 was investigated as signals related to cytoskeleton dynamics. The polymerization of GFAP and the expression of focal contact structured by vinculin were found to be altered, especially after 48 and 72 h of microgravity. Connexin-43, involved in several intracellular pathways that critically promote cell motility and invasion of glioma cells, was found to be largely reduced following microgravity exposure. In conclusion, microgravity, by reducing the expression of Connexin-43, alters the architecture of specific cytoskeletal elements such as GFAP and increases the focal contact, which can induce a reduction in glioma cell mobility, thereby inhibiting their aggressive metastatic behavior.
Full article
(This article belongs to the Special Issue State-of-the-Art Biophysics in Italy)
►▼
Show Figures
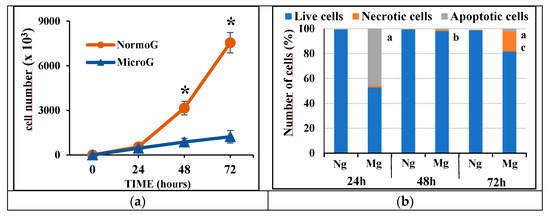
Figure 1
Open AccessArticle
Investigating the Insertion Mechanism of Cell-Penetrating Peptide Penetratin into Cell Membranes: Implications for Targeted Drug Delivery
by
Bashiyar Almarwani, Yahia Z. Hamada, Nsoki Phambu and Anderson Sunda-Meya
Biophysica 2023, 3(4), 620-635; https://doi.org/10.3390/biophysica3040042 - 11 Nov 2023
Abstract
The cell-penetrating peptide (CPP) penetratin (PEN) has garnered attention for its potential to enter tumor cells. However, its translocation mechanism and lack of selectivity remain debated. This study investigated PEN’s insertion into healthy cells (H-) and cancer cells (C-) using micromolar concentrations and
[...] Read more.
The cell-penetrating peptide (CPP) penetratin (PEN) has garnered attention for its potential to enter tumor cells. However, its translocation mechanism and lack of selectivity remain debated. This study investigated PEN’s insertion into healthy cells (H-) and cancer cells (C-) using micromolar concentrations and various techniques. Raman spectroscopy was used to determine PEN’s location in the lipid bilayer at different lipid-to-peptide ratios. Dynamic light scattering (DLS) and zeta potential analysis were used to measure the lipid–PEN complex’s size and charge. The results showed helical PEN particles directly inserted into C- membranes at a ratio of 110, while aggregated particles stayed on H- surfaces. Raman spectroscopy and scanning electron microscopy confirmed PEN insertion in C- membranes. Zeta potential studies revealed highly negative charges for PEN–C- complexes and neutral charges for PEN–H- complexes at pH 6.8. C- integrity remained unchanged at a ratio of 110. Specific lipid-to-peptide ratios with dipalmitoylphosphatidylserine (DPPS) were crucial for direct insertion. These results provide valuable insights into CPP efficacy for targeted drug delivery in cancer cells, considering membrane composition and lipid-to-peptide ratios.
Full article
(This article belongs to the Special Issue Biomedical Optics 2.0)
►▼
Show Figures
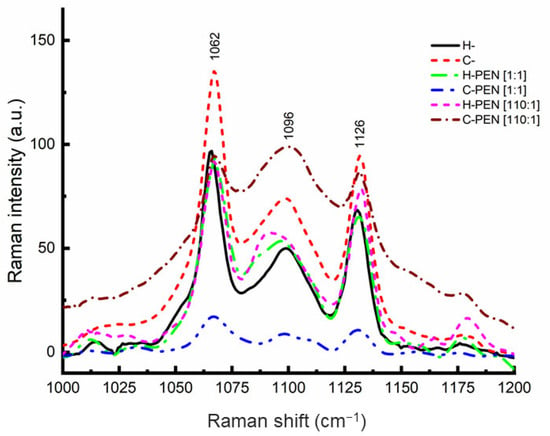
Figure 1
Open AccessReview
Physical Virology in Spain
by
David Reguera, Pedro J. de Pablo, Nicola G. A. Abrescia, Mauricio G. Mateu, Javier Hernández-Rojas, José R. Castón and Carmen San Martín
Biophysica 2023, 3(4), 598-619; https://doi.org/10.3390/biophysica3040041 - 31 Oct 2023
Abstract
Virus particles consist of a protein coat that protects their genetic material and delivers it to the host cell for self-replication. Understanding the interplay between virus structure and function is a requirement for understanding critical processes in the infectious cycle such as entry,
[...] Read more.
Virus particles consist of a protein coat that protects their genetic material and delivers it to the host cell for self-replication. Understanding the interplay between virus structure and function is a requirement for understanding critical processes in the infectious cycle such as entry, uncoating, genome metabolism, capsid assembly, maturation, and propagation. Together with well-established techniques in cell and molecular biology, physical virology has emerged as a rapidly developing field, providing detailed, novel information on the basic principles of virus assembly, disassembly, and dynamics. The Spanish research community contains a good number of groups that apply their knowledge on biology, physics, or chemistry to the study of viruses. Some of these groups got together in 2010 under the umbrella of the Spanish Interdisciplinary Network on Virus Biophysics (BioFiViNet). Thirteen years later, the network remains a fertile ground for interdisciplinary collaborations geared to reveal new aspects on the physical properties of virus particles, their role in regulating the infectious cycle, and their exploitation for the development of virus-based nanotechnology tools. Here, we highlight some achievements of Spanish groups in the field of physical virology.
Full article
(This article belongs to the Special Issue State-of-the-Art Biophysics in Spain 2.0)
►▼
Show Figures
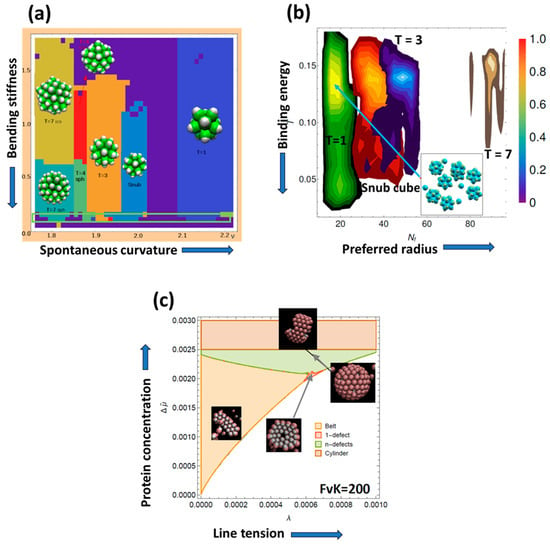
Figure 1
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
Special Issues
Special Issue in
Biophysica
Protein Disorder
Guest Editor: Beata Greb-MarkiewiczDeadline: 31 May 2024
Special Issue in
Biophysica
The Structure and Function of Proteins, Lipids, and Nucleic Acids
Guest Editor: Ivo CrnolatacDeadline: 30 June 2024
Special Issue in
Biophysica
Biophysical Studies of Metalloproteins
Guest Editor: Maria Carmela Bonaccorsi Di PattiDeadline: 28 August 2024
Special Issue in
Biophysica
Live Cell Microscopy
Guest Editors: Herbert Schneckenburger, Christoph CremerDeadline: 30 September 2024
Topical Collections
Topical Collection in
Biophysica
Feature Papers in Biophysics
Collection Editors: Ricardo L. Mancera, Paul C. Whitford, Chandra Kothapalli