Metabolomics and Integrated Multi-Omics in Health and Disease
A topical collection in Biomolecules (ISSN 2218-273X). This collection belongs to the section "Molecular Medicine".
Viewed by 94965Editors
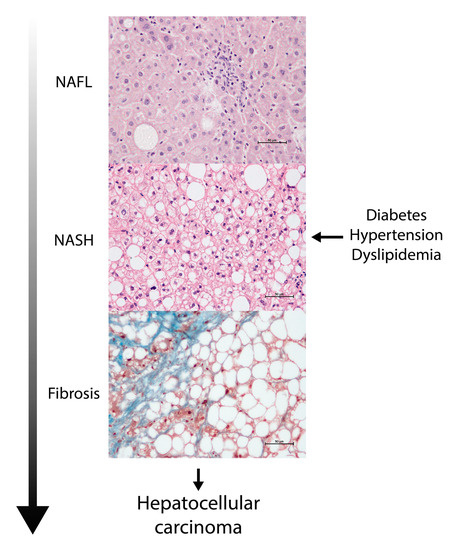
Interests: obesity-related metabolic complications; diabetes; non-alcoholic fatty liver disease; cancer; arteriosclerosis; metabolism; metabolomics; lipidomics; epigenetics; oxidation; inflammation; metformin
Special Issues, Collections and Topics in MDPI journals
Interests: polyphenols; metabolomics, chromatography mass spectrometry; analytical method development; bioactivity; cell culture; gas chromatography–mass spectrometry; cell biology; biostatistics
Interests: inflammation; macrophage; case-control studies; insulin signaling; energy metabolism; insulin resistance; cell culture; cytokines; metabolism; glucose metabolism
Interests: gene regulation; signal transduction; mass spectrometry methods; epigenetics; LC-MS LC-MS/MS; high throughput sequencing; genetic analysis affinity chromatography
Topical Collection Information
Dear Colleagues,
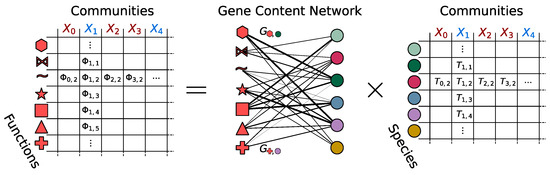
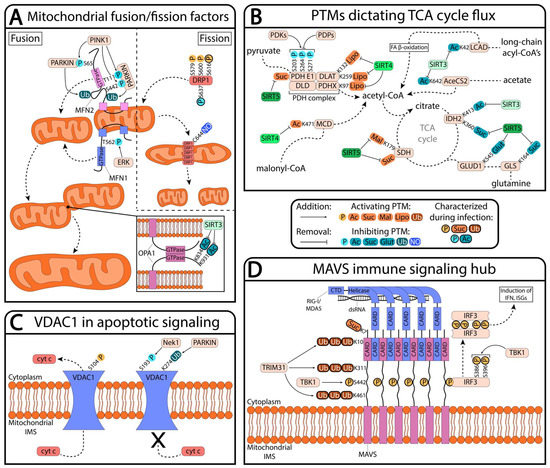
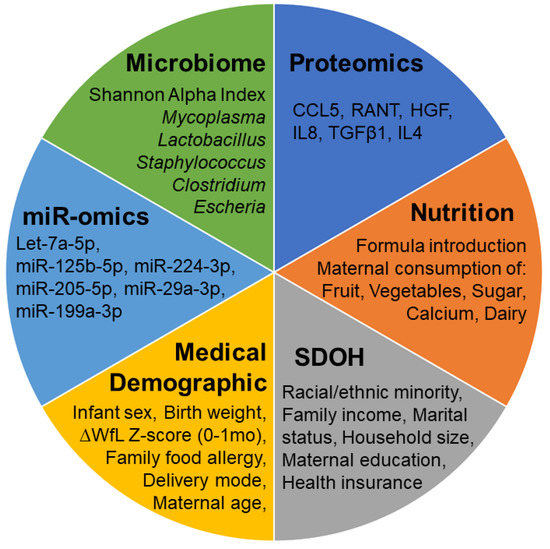
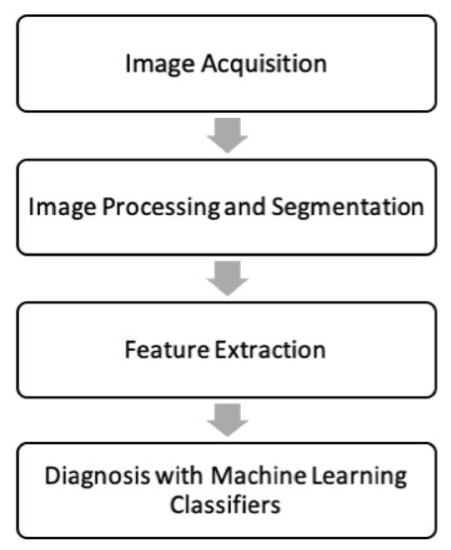
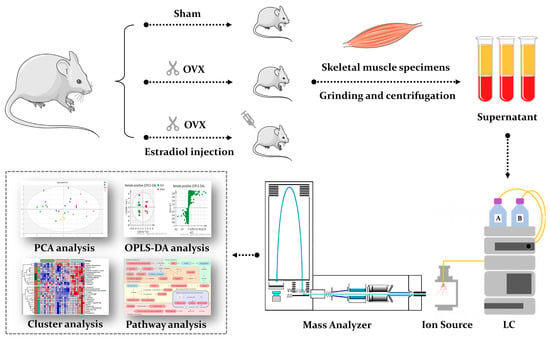
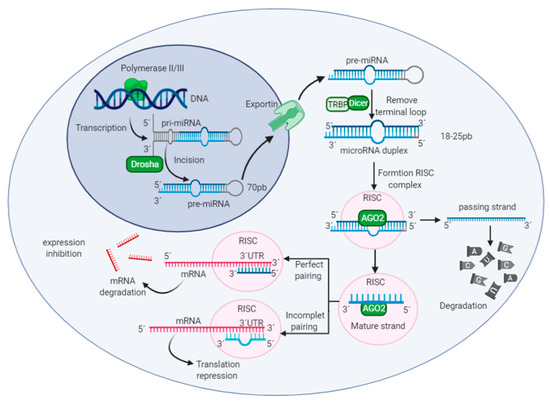
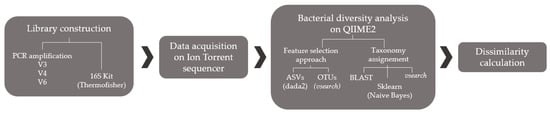
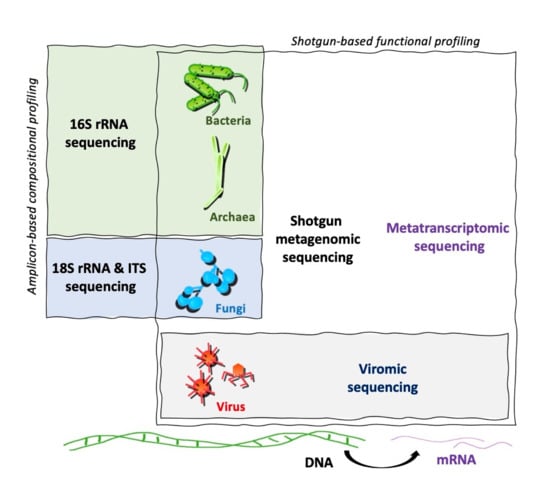
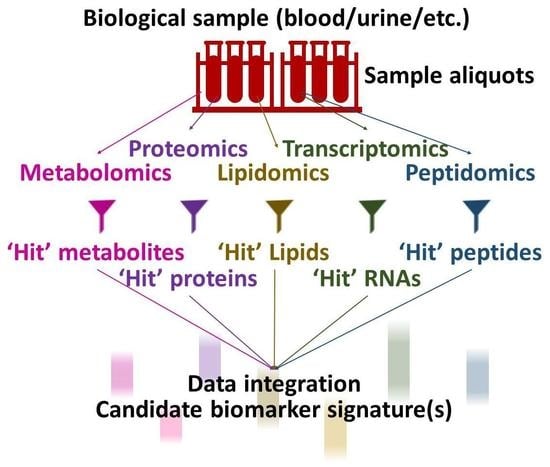
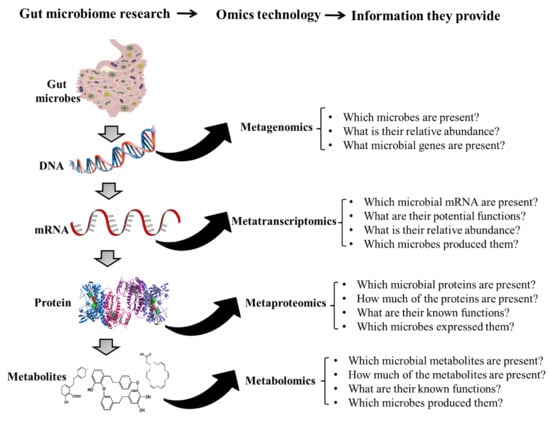
Metabolomics and the integration of multi-omics have emerged as key platforms for the understanding of healthy status and the association of metabolic disturbances with diseases. Much progress has been made in the last decade to obtain accurate metabolic profiles, and it is now standard practice. Both nuclear magnetic resonance and mass spectrometry are effective tools that analyze the molecular composition of a sample. Nevertheless, challenges remain in multiple clinical and methodological areas. There are virtually no fields in medicine in which the correct approaches do not result in possible new biologic insights based on adequate designs, instrumentation, and methods and, in particular, software for correct interpretation and data mining. Applications are not limited to “classical” metabolic diseases but also have a wide range of uses in drug discovery, cancer, cardiovascular health, aging, immunology, epigenetics or microbiome. Finding compounds, derived from natural sources, with the ability to interfere in a disease state and promote health is currently a hot topic. More recent analysis has provided evidence that some of the metabolome variation observed across individuals may be directly encoded in the genome, which might be valuable in personalized medicine. Therefore, this Topical Collection of Biomolecules welcomes all submissions of manuscripts, original research, comprehensive reviews, personal opinion focused on the fields and methods outlined. The following keywords describe the immediate interest and expertise from the Editors.
Prof. Jorge Joven
Dr. Fernández-Arroyo Salvador
Dr. Anna Hernández-Aguilera
Dr. Nuria Canela
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomolecules is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- metabolomics
- lipidomics
- proteomics
- biomarker discovery and validation
- computational tools
- food science
- obesity
- diabetes