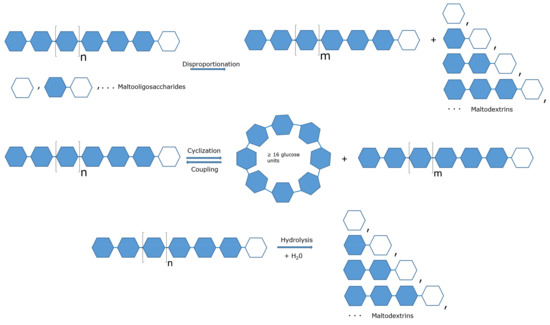
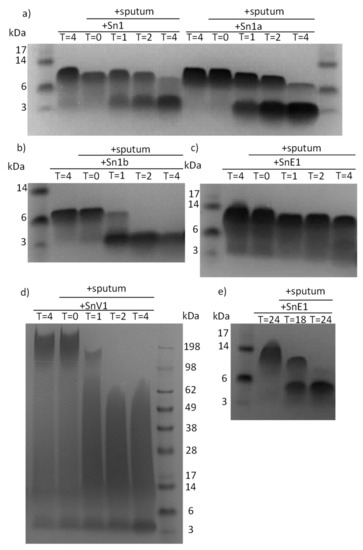
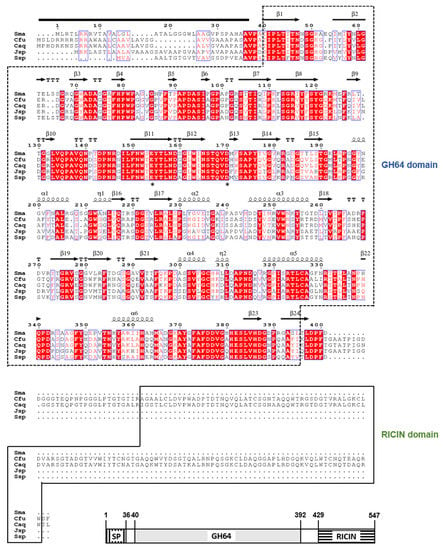
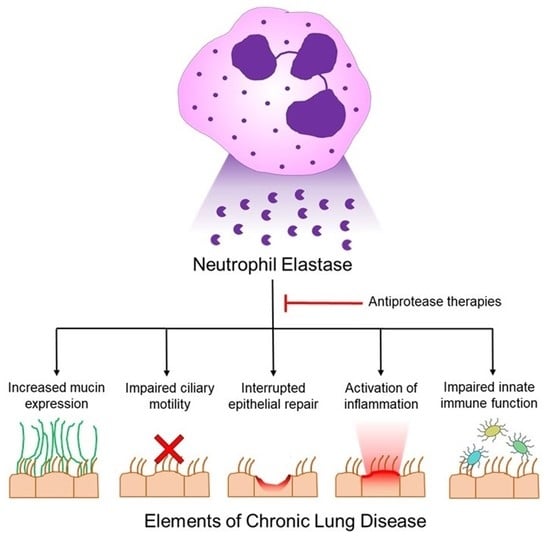
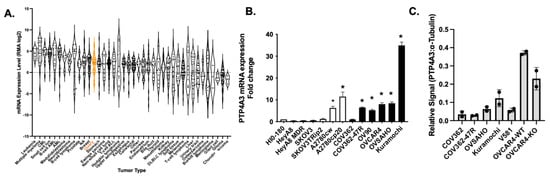
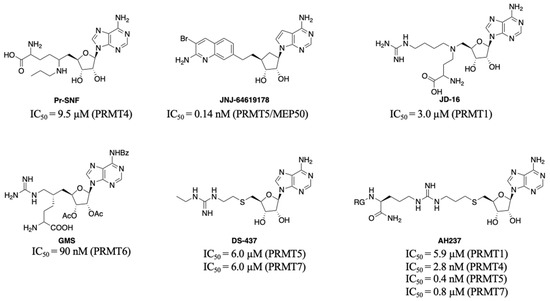
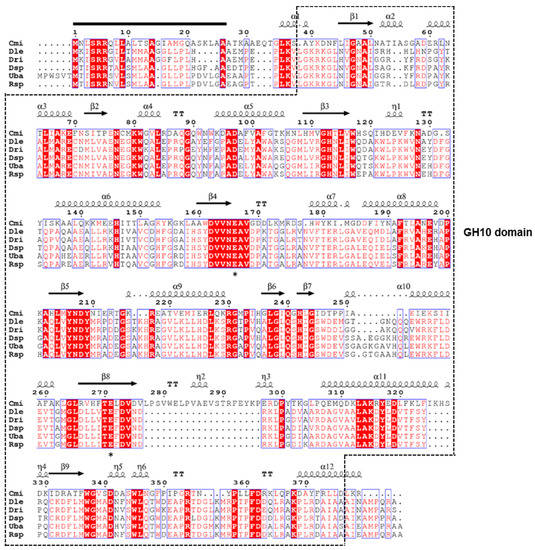
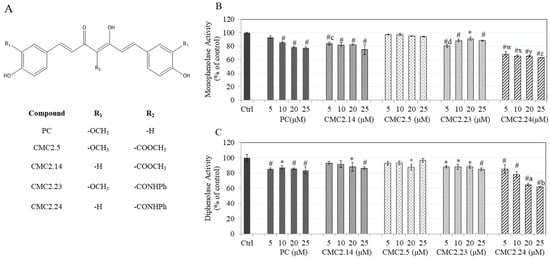
Feature Papers in Enzymology (Closed)
A topical collection in Biomolecules (ISSN 2218-273X). This collection belongs to the section "Enzymology".
Viewed by 54411Editor
Interests: drug discovery; chemical biology; biological macromolecules; glycosaminoglycans; coagulation factors; cancer; viral infection; bio-mimetic design; enzyme mechanisms; computational biology; high throughput screening
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Topical Collection entitled “Feature Papers in Enzymology” aims to collect high-quality research articles, review articles, and communications on all aspects of enzymology. It is dedicated to recent advances in the broad research area of enzymology; is inclusive of papers focused on structure, function, and properties of enzymes as well as catalytic and inhibition mechanisms of enzymes, mechanisms of drug action, and related advances on enzymes. The collection will highlight advances in all types of enzymes including hydrolases, oxidoreductases, lyases, transferases, ligases, and isomerases. Research related to cofactors (prosthetic groups, coenzymes and metal ions) will also be of interest to the collection. It will be comprised of a selection of exclusive papers from the Editorial Board Members (EBMs) of the Enzymology Section, as well as invited papers from relevant experts. We also welcome senior experts in the field to make contributions to this Topical Collection. We aim to represent our section as an attractive open-access publishing platform for the enzymology research field.
Prof. Dr. Umesh Desai
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomolecules is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- structure and function
- enzyme catalysis
- enzyme inhibition
- enzyme mechanisms
- computational enzymology
- biophysical characterization
- structure determination
- drug action
- biochemical mechanisms
- molecular forces
- allosterism
- orthosterism