DNA Repair and Immune Response
Share This Topical Collection
Editor
 Dr. Valentyn Oksenych
Dr. Valentyn Oksenych
 Dr. Valentyn Oksenych
Dr. Valentyn Oksenych
E-Mail
Website
Collection Editor
Broegelmann Research Laboratory, Department of Clinical Science, University of Bergen, 5020 Bergen, Norway
Interests: DNA repair; DNA damage response; genetics; primary immunodeficiency; B lymphocyte development; mouse models
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Lesions are generated continuously in our cellular DNA by external and internal factors. DNA damage is recognized and repaired by several pathways that involve multiple proteins. DNA damage response (DDR) is a complex change in cellular metabolism following DNA damage. Failures in DNA repair and DDR result in developmental disorders, defects in immune response, abnormal neurodevelopment, cancer, and aging. We invite the submission of original research manuscripts and review articles that cover different aspects of DNA repair, DDR, and related topics, such as immune response, neurodevelopment, and cancer. I am looking forward to your contribution.
Dr. Valentyn Oksenych
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomolecules is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- DNA damage response
- DNA repair
- cell death
- genomic instability
- innate immunity
- adaptive immunity
- cancer
- neurodevelopment
Published Papers (8 papers)
Open AccessReview
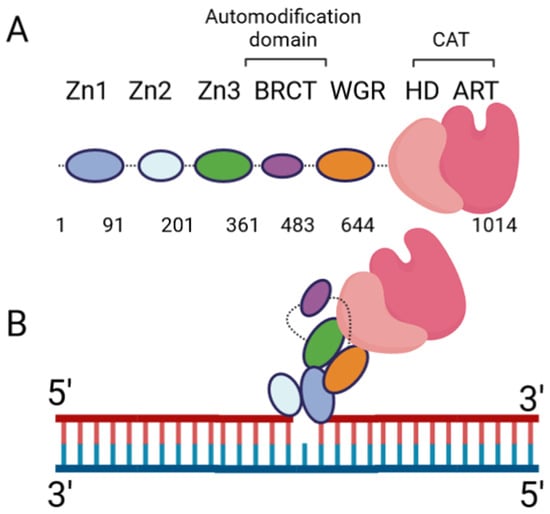
The Role of Poly(ADP-ribose) Polymerase 1 in Nuclear and Mitochondrial Base Excision Repair
by
Geoffrey K. Herrmann and Y. Whitney Yin
Cited by 4 | Viewed by 1748
Abstract
Poly(ADP-ribose) (PAR) Polymerase 1 (PARP-1), also known as ADP-ribosyl transferase with diphtheria toxin homology 1 (ARTD-1), is a critical player in DNA damage repair, during which it catalyzes the ADP ribosylation of self and target enzymes. While the nuclear localization of PARP-1 has
[...] Read more.
Poly(ADP-ribose) (PAR) Polymerase 1 (PARP-1), also known as ADP-ribosyl transferase with diphtheria toxin homology 1 (ARTD-1), is a critical player in DNA damage repair, during which it catalyzes the ADP ribosylation of self and target enzymes. While the nuclear localization of PARP-1 has been well established, recent studies also suggest its mitochondrial localization. In this review, we summarize the differences between mitochondrial and nuclear Base Excision Repair (BER) pathways, the involvement of PARP-1 in mitochondrial and nuclear BER, and its functional interplay with other BER enzymes.
Full article
►▼
Show Figures
Open AccessArticle
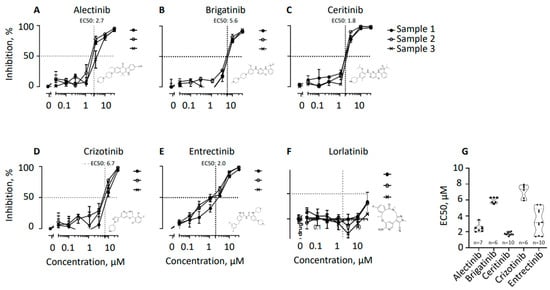
Tyrosine Kinase Inhibitors Target B Lymphocytes
by
Nikki Lyn Esnardo Upfold, Pavlo Petakh, Aleksandr Kamyshnyi and Valentyn Oksenych
Viewed by 2104
Abstract
Autoimmune disorders and some types of blood cancer originate when B lymphocytes malfunction. In particular, when B cells produce antibodies recognizing the body’s proteins, it leads to various autoimmune disorders. Additionally, when B cells of various developmental stages transform into cancer cells, it
[...] Read more.
Autoimmune disorders and some types of blood cancer originate when B lymphocytes malfunction. In particular, when B cells produce antibodies recognizing the body’s proteins, it leads to various autoimmune disorders. Additionally, when B cells of various developmental stages transform into cancer cells, it results in blood cancers, including multiple myeloma, lymphoma, and leukemia. Thus, new methods of targeting B cells are required for various patient groups. Here, we used protein kinase inhibitors alectinib, brigatinib, ceritinib, crizotinib, entrectinib, and lorlatinib previously approved as drugs treating anaplastic lymphoma kinase (ALK)-positive lung cancer cells. We hypothesized that the same inhibitors will efficiently target leukocyte tyrosine kinase (LTK)-positive, actively protein-secreting mature B lymphocytes, including plasma cells. We isolated CD19-positive human B cells from the blood of healthy donors and used two alternative methods to stimulate cell maturation toward plasma cells. Using cell proliferation and flow cytometry assays, we found that ceritinib and entrectinib eliminate plasma cells from B cell populations. Alectinib, brigatinib, and crizotinib also inhibited B cell proliferation, while lorlatinib had no or limited effect on B cells. More generally, we concluded that several drugs previously developed to treat ALK-positive malignant cells can be also used to treat LTK-positive B cells.
Full article
►▼
Show Figures
Open AccessEditorial
DNA Repair and Immune Response: Editorial
by
Valentyn Oksenych
Cited by 2 | Viewed by 1639
Abstract
Developing B and T lymphocytes requires programmed DNA double-strand breaks followed by the activation of the DNA damage response (DDR) pathway and DNA repair [...]
Full article
Open AccessFeature PaperReview
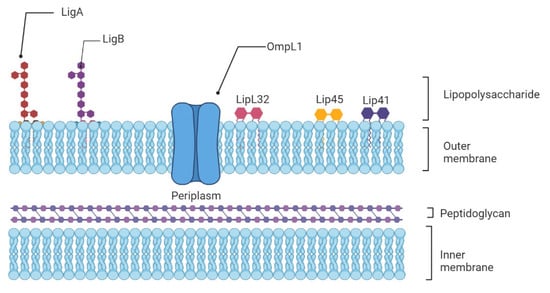
Weil’s Disease—Immunopathogenesis, Multiple Organ Failure, and Potential Role of Gut Microbiota
by
Pavlo Petakh, Vitaliia Isevych, Aleksandr Kamyshnyi and Valentyn Oksenych
Cited by 9 | Viewed by 5141
Abstract
Leptospirosis is an important zoonotic disease, causing about 60,000 deaths annually. In this review, we have described in detail the immunopathogenesis of leptospirosis, the influence of cytokines, genetic susceptibility on the course of the disease, and the evasion of the immune response. These
[...] Read more.
Leptospirosis is an important zoonotic disease, causing about 60,000 deaths annually. In this review, we have described in detail the immunopathogenesis of leptospirosis, the influence of cytokines, genetic susceptibility on the course of the disease, and the evasion of the immune response. These data are combined with information about immunological and pathomorphological changes in the kidneys, liver, and lungs, which are most affected by Weil’s disease. The review also suggests a possible role of the gut microbiota in the clinical course of leptospirosis, the main mechanisms of the influence of gut dysbiosis on damage in the liver, kidneys, and lungs through several axes, i.e., gut-liver, gut-kidney, and gut-lungs. Modulation of gut microbiota by probiotics and/or fecal microbiota transplantation in leptospirosis may become an important area of scientific research.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceArticle
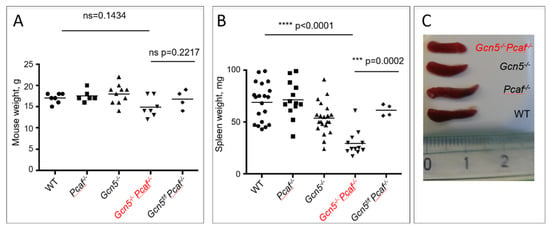
Acetyltransferases GCN5 and PCAF Are Required for B Lymphocyte Maturation in Mice
by
Valentyn Oksenych, Dan Su and Jeremy A. Daniel
Cited by 4 | Viewed by 2638
Abstract
B lymphocyte development has two DNA recombination processes: V(D)J recombination of the immunoglobulin (
Igh) gene variable region, and class switching of the
Igh constant regions from IgM to IgG, IgA, or IgE. V(D)J recombination is required for the successful maturation of
[...] Read more.
B lymphocyte development has two DNA recombination processes: V(D)J recombination of the immunoglobulin (
Igh) gene variable region, and class switching of the
Igh constant regions from IgM to IgG, IgA, or IgE. V(D)J recombination is required for the successful maturation of B cells from pro-B to pre-B to immature-B and then to mature B cells in the bone marrow. CSR occurs outside of the bone marrow when mature B cells migrate to peripheral lymphoid organs, such as spleen and lymph nodes. Both V(D)J recombination and CSR depend on an open chromatin state that makes DNA accessible to specific enzymes, recombination activating gene (RAG), and activation-induced cytidine deaminase (AID). Acetyltransferases GCN5 and PCAF possess redundant functions acetylating histone H3 lysine 9 (H3K9). Here, we generated a mouse model that lacked both GCN5 and PCAF in B cells. Double-deficient mice possessed low levels of mature B cells in the bone marrow and peripheral organs, an accumulation of pro-B cells in bone marrow, and reduced CSR levels. We concluded that both GCN5 and PCAF are required for B-cell development in vivo.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceReview
DNA End Joining: G0-ing to the Core
by
Richard L. Frock, Cheyenne Sadeghi, Jodie Meng and Jing L. Wang
Cited by 9 | Viewed by 4376
Abstract
Humans have evolved a series of DNA double-strand break (DSB) repair pathways to efficiently and accurately rejoin nascently formed pairs of double-stranded DNA ends (DSEs). In G0/G1-phase cells, non-homologous end joining (NHEJ) and alternative end joining (A-EJ) operate to support covalent rejoining of
[...] Read more.
Humans have evolved a series of DNA double-strand break (DSB) repair pathways to efficiently and accurately rejoin nascently formed pairs of double-stranded DNA ends (DSEs). In G0/G1-phase cells, non-homologous end joining (NHEJ) and alternative end joining (A-EJ) operate to support covalent rejoining of DSEs. While NHEJ is predominantly utilized and collaborates extensively with the DNA damage response (DDR) to support pairing of DSEs, much less is known about A-EJ collaboration with DDR factors when NHEJ is absent. Non-cycling lymphocyte progenitor cells use NHEJ to complete V(D)J recombination of antigen receptor genes, initiated by the RAG1/2 endonuclease which holds its pair of targeted DSBs in a synapse until each specified pair of DSEs is handed off to the NHEJ DSB sensor complex, Ku. Similar to designer endonuclease DSBs, the absence of Ku allows for A-EJ to access RAG1/2 DSEs but with random pairing to complete their repair. Here, we describe recent insights into the major phases of DSB end joining, with an emphasis on synapsis and tethering mechanisms, and bring together new and old concepts of NHEJ vs. A-EJ and on RAG2-mediated repair pathway choice.
Full article
►▼
Show Figures
Open AccessArticle
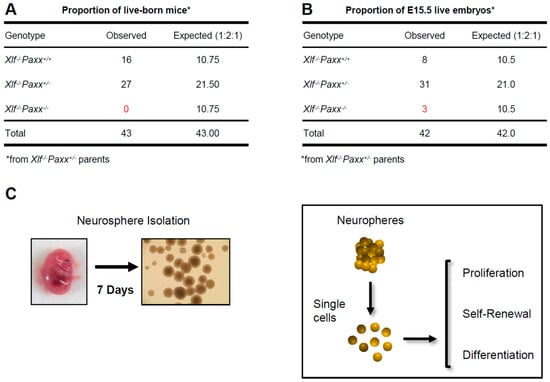
Non-Homologous End Joining Factors XLF, PAXX and DNA-PKcs Maintain the Neural Stem and Progenitor Cell Population
by
Raquel Gago-Fuentes and Valentyn Oksenych
Cited by 4 | Viewed by 2815
Abstract
Non-homologous end-joining (NHEJ) is a major DNA repair pathway in mammalian cells that recognizes, processes and fixes DNA damage throughout the cell cycle and is specifically important for homeostasis of post-mitotic neurons and developing lymphocytes. Neuronal apoptosis increases in the mice lacking NHEJ
[...] Read more.
Non-homologous end-joining (NHEJ) is a major DNA repair pathway in mammalian cells that recognizes, processes and fixes DNA damage throughout the cell cycle and is specifically important for homeostasis of post-mitotic neurons and developing lymphocytes. Neuronal apoptosis increases in the mice lacking NHEJ factors Ku70 and Ku80. Inactivation of other NHEJ genes, either
Xrcc4 or
Lig4, leads to massive neuronal apoptosis in the central nervous system (CNS) that correlates with embryonic lethality in mice. Inactivation of either
Paxx,
Mri or
Dna-pkcs NHEJ gene results in normal CNS development due to compensatory effects of
Xlf. Combined inactivation of
Xlf/Paxx,
Xlf/Mri and
Xlf/Dna-pkcs, however, results in late embryonic lethality and high levels of apoptosis in CNS. To determine the impact of NHEJ factors on the early stages of neurodevelopment, we isolated neural stem and progenitor cells from mouse embryos and investigated proliferation, self-renewal and differentiation capacity of these cells lacking either
Xlf,
Paxx,
Dna-pkcs,
Xlf/Paxx or
Xlf/Dna-pkcs. We found that XRCC4-like factor (XLF), DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and paralogue of XRCC4 and XLF (PAXX) maintain the neural stem and progenitor cell populations and neurodevelopment in mammals, which is particularly evident in the double knockout models.
Full article
►▼
Show Figures
Open AccessArticle
Development of Methods Derived from Iodine-Induced Specific Cleavage for Identification and Quantitation of DNA Phosphorothioate Modifications
by
Sucheng Zhu, Tao Zheng, Lingxin Kong, Jinli Li, Bo Cao, Michael S. DeMott, Yihua Sun, Ying Chen, Zixin Deng, Peter C. Dedon and Delin You
Cited by 4 | Viewed by 2433
Abstract
DNA phosphorothioate (PT) modification is a novel modification that occurs on the DNA backbone, which refers to a non-bridging phosphate oxygen replaced by sulfur. This exclusive DNA modification widely distributes in bacteria but has not been found in eukaryotes to date. PT modification
[...] Read more.
DNA phosphorothioate (PT) modification is a novel modification that occurs on the DNA backbone, which refers to a non-bridging phosphate oxygen replaced by sulfur. This exclusive DNA modification widely distributes in bacteria but has not been found in eukaryotes to date. PT modification renders DNA nuclease tolerance and serves as a constitute element of bacterial restriction–modification (R–M) defensive system and more biological functions are awaiting exploration. Identification and quantification of the bacterial PT modifications are thus critical to better understanding their biological functions. This work describes three detailed methods derived from iodine-induced specific cleavage-an iodine-induced cleavage assay (ICA), a deep sequencing of iodine-induced cleavage at PT site (ICDS) and an iodine-induced cleavage PT sequencing (PT-IC-Seq)-for the investigation of PT modifications. Using these approaches, we have identified the presence of PT modifications and quantized the frequency of PT modifications in bacteria. These characterizations contributed to the high-resolution genomic mapping of PT modifications, in which the distribution of PT modification sites on the genome was marked accurately and the frequency of the specific modified sites was reliably obtained. Here, we provide time-saving and less labor-consuming methods for both of qualitative and quantitative analysis of genomic PT modifications. The application of these methodologies will offer great potential for better understanding the biology of the PT modifications and open the door to future further systematical study.
Full article
►▼
Show Figures