Journal Description
Bacteria
Bacteria
is an international, peer-reviewed, open access journal on bacteriology published quarterly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- Rapid Publication: first decisions in 16 days; acceptance to publication in 5.8 days (median values for MDPI journals in the second half of 2023).
- Recognition of Reviewers: APC discount vouchers, optional signed peer review, and reviewer names published annually in the journal.
- Bacteria is a companion journal of Pathogens.
Latest Articles
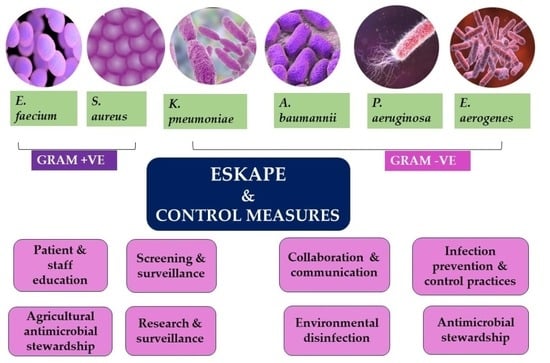
ESKAPE: Navigating the Global Battlefield for Antimicrobial Resistance and Defense in Hospitals
Bacteria 2024, 3(2), 76-98; https://doi.org/10.3390/bacteria3020006 - 16 Apr 2024
Abstract
►
Show Figures
The current healthcare environment is at risk due to the facilitated transmission and empowerment of the ESKAPE pathogens, comprising of Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species. These pathogens have posed significant
[...] Read more.
The current healthcare environment is at risk due to the facilitated transmission and empowerment of the ESKAPE pathogens, comprising of Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species. These pathogens have posed significant challenges to global public health and the threat has only amplified over time. These multidrug-resistant bacteria have become adept at escaping the effects of conventional antibiotics utilized, leading to severe healthcare-associated infections and compromising immunocompromised patient outcomes to a greater extent. The impact of ESKAPE pathogens is evident in the rapidly rising rates of treatment failures, increased mortality, and elevated healthcare costs. To combat this looming crisis, diverse strategies have been adopted, ranging from the development of novel antimicrobial agents and combination therapies to the implementation of stringent infection control measures. Additionally, there has been a growing emphasis on promoting antimicrobial stewardship programs to optimize the use of existing antibiotics and reduce the selective pressure driving the evolution of resistance. While progress has been made to some extent, the rapid adaptability of these pathogens and the enhancement of antimicrobial resistance mechanisms proves to be a major hurdle yet to be crossed by healthcare professionals. In this viewpoint, the impending threat heralded by the proliferation of ESKAPE pathogens, and the need for a concerted global effort via international collaborations for the assurance of effective and sustainable solutions, are explored. To curb the possibility of outbreaks in the future and to safeguard public health, better preparation via global awareness and defense mechanisms should be given paramount importance.
Full article
Open AccessReview
Role of Plant Growth Promoting Rhizobacteria (PGPR) as a Plant Growth Enhancer for Sustainable Agriculture: A Review
by
Asma Hasan, Baby Tabassum, Mohammad Hashim and Nagma Khan
Bacteria 2024, 3(2), 59-75; https://doi.org/10.3390/bacteria3020005 - 01 Apr 2024
Abstract
The rhizosphere of a plant is home to helpful microorganisms called plant growth-promoting rhizobacteria (PGPR), which play a crucial role in promoting plant growth and development. The significance of PGPR for long-term agricultural viability is outlined in this review. Plant growth processes such
[...] Read more.
The rhizosphere of a plant is home to helpful microorganisms called plant growth-promoting rhizobacteria (PGPR), which play a crucial role in promoting plant growth and development. The significance of PGPR for long-term agricultural viability is outlined in this review. Plant growth processes such as nitrogen fixation, phosphate solubilization, and hormone secretion are discussed. Increased plant tolerance to biotic and abiotic stress, reduced use of chemical fertilizers and pesticides, and enhanced nutrient availability, soil fertility, and absorption are all mentioned as potential benefits of PGPR. PGPR has multiple ecological and practical functions in the soil’s rhizosphere. One of PGPR’s various roles in agroecosystems is to increase the synthesis of phytohormones and other metabolites, which have a direct impact on plant growth. Phytopathogens can be stopped in their tracks, a plant’s natural defenses can be bolstered, and so on. PGPR also helps clean up the soil through a process called bioremediation. The PGPR’s many functions include indole acetic acid (IAA) production, ammonia (NH3) production, hydrogen cyanide (HCN) production, catalase production, and more. In addition to aiding in nutrient uptake, PGPR controls the production of a hormone that increases root size and strength. Improving crop yield, decreasing environmental pollution, and guaranteeing food security are only some of the ecological and economic benefits of employing PGPR for sustainable agriculture.
Full article
(This article belongs to the Special Issue Interaction between Plants and Growth-Promoting Rhizobacteria (PGPR) for Sustainable Development)
►▼
Show Figures
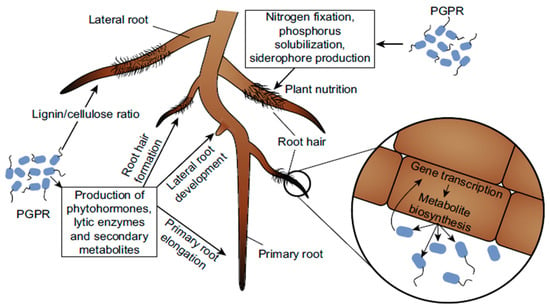
Figure 1
Open AccessArticle
Streptococcus thermophilus and Lactobacillus bulgaricus Attributes as Influenced by Carao (Cassia grandis) Fruit Parts
by
Miguel Mariano Tabora, Ricardo S. Aleman, Ashley Castro, Allan Avila, Dany Avila, David Picha, Roberto Cedillos, Shirin Kazemzadeh, Leyla K. Pournaki, Ajitesh Yaday, Jhunior Marcia and Aryana Kayanush
Bacteria 2024, 3(2), 42-58; https://doi.org/10.3390/bacteria3020004 - 30 Mar 2024
Abstract
►▼
Show Figures
Carao (Cassia grandis) contains numerous bioactive substances that contribute to gastrointestinal well-being. The present study assessed the potential impacts of carao on the viability and performance of Streptococcus thermophilus and Lactobacillus bulgaricus under various adverse conditions. These conditions included bile, acid,
[...] Read more.
Carao (Cassia grandis) contains numerous bioactive substances that contribute to gastrointestinal well-being. The present study assessed the potential impacts of carao on the viability and performance of Streptococcus thermophilus and Lactobacillus bulgaricus under various adverse conditions. These conditions included bile, acid, gastric juice, and lysozyme exposure, simulating the digestive process from the mouth to the intestines. The activity of proteases from cultures was monitored to examine their proteolytic capabilities. To achieve this, the cultures were cultivated in a solution containing plant material, and the results were compared against a control sample after an incubation period. Subsequently, the total phenolic content, total carotenoid content, antioxidant activity, sugar profile, and acid profile of the plant materials were analyzed. These analyses were conducted to explore these compounds’ influence on cultures’ survival. Seeds contained the highest total phenols (766.87 ± 11.56 µg GAE/mL), total carotenoid content (7.43 ± 0.31 mg Q/mL), and antioxidant activity (40.76 ± 1.87%). Pulp contained the highest moisture (12.55 ± 0.44%), ash (6.45 ± 0.15%), lipid (0.66 ± 0.07%), protein (16.56 ± 0.21%), sucrose (9.07 ± 0.78 g/100 g), and fructose (3.76 ± 0.06 g/ 100 g). The crust had the highest content of ash (85.14 ± 0.27%) and succinic acid (2.01 ± 0.06 g/100 g). Results indicated that seeds negatively affected cultures’ survival in the bile tolerance test and had positive effects on Lactobacillus bulgaricus in the protease activity test. Otherwise, the other carao tissues could not change the results significantly (p > 0.05) compared to the control in different tests. The carao crust positively affected cultures’ against protease activity, especially in Lactobacillus bulgaricus, and had a negative effect on the growth of S. thermophilus in the lysozyme and gastric acid resistance test.
Full article
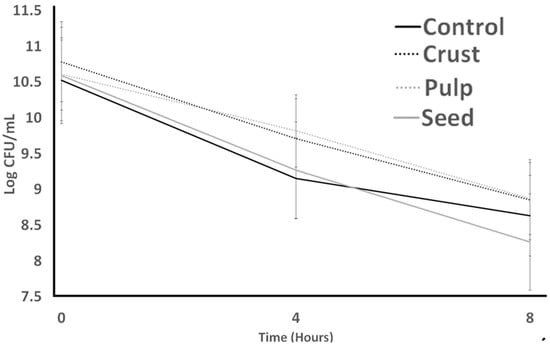
Figure 1
Open AccessArticle
Impact of Novel Functional Ingredients on Lactobacillus casei Viability
by
Ricardo S. Aleman, Franklin Delarca, Mallerly Sarmientos, Jhunior Marcia, Ajitesh Yaday and Aryana Kayanush
Bacteria 2024, 3(1), 30-41; https://doi.org/10.3390/bacteria3010003 - 19 Mar 2024
Abstract
►▼
Show Figures
Nipple fruit (Solanum mammosum), teosinte (Dioon mejiae), Caesar mushroom (Amanita caesarea), and weevil (Rhynchophorus palmarum) powders have shown great nutritional content with meaningful dietary applications. This study aspired to investigate the impact of nipple fruit, teosinte,
[...] Read more.
Nipple fruit (Solanum mammosum), teosinte (Dioon mejiae), Caesar mushroom (Amanita caesarea), and weevil (Rhynchophorus palmarum) powders have shown great nutritional content with meaningful dietary applications. This study aspired to investigate the impact of nipple fruit, teosinte, Caesar mushroom, and weevil powders on the bile tolerance, acid tolerance, lysozyme tolerance, gastric juice resistance, and protease activity of Lactobacillus casei. Nipple fruit, teosinte, Caesar mushroom, and weevil powders were combined at 2% (wt/vol), whereas the control samples did not include the ingredients. The bile and acid tolerances were analyzed in Difco De Man–Rogosa–Sharpe broth incubated under aerobic conditions at 37 °C. The bile tolerance was investigated by adding 0.3% oxgall, whereas the acid tolerance was studied by modifying the pH to 2.0. The lysozyme tolerance was studied in electrolyte solution containing lysozyme (100 mg/L), while the gastric juice tolerance was analyzed at pH levels of 2, 3, 4, 5, and 7. The protease activity was studied spectrophotometrically at 340 nm in skim milk incubated under aerobic conditions at 37 °C. The results show that nipple fruit increased the counts, whereas Caesar mushroom and weevil powders resulted in lower counts for bile tolerance, acid tolerance, lysozyme resistance, and simulated gastric juice tolerance characteristics. Furthermore, the protease activity increased by adding nipple fruit to skim milk. According to the results, nipple fruit may improve the characteristics of L. casei in cultured dairy by-products.
Full article
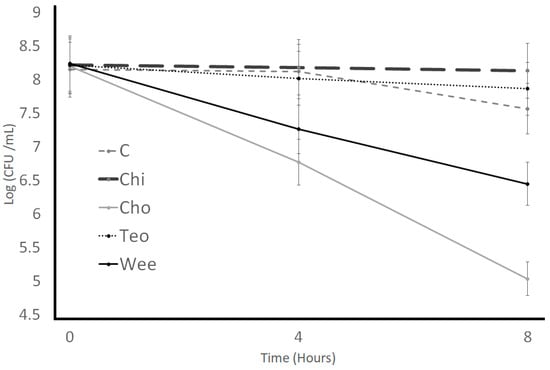
Figure 1
Open AccessReview
Harnessing the Power of Zinc-Solubilizing Bacteria: A Catalyst for a Sustainable Agrosystem
by
Swapnil Singh, Rohit Chhabra, Ashish Sharma and Aditi Bisht
Bacteria 2024, 3(1), 15-29; https://doi.org/10.3390/bacteria3010002 - 24 Feb 2024
Abstract
A variety of agrochemicals, especially fertilizers, are applied indiscriminately by farmers across trapezoidal landscapes to increase productivity and satisfy the rising food demand. Around one-third of the populace in developing nations is susceptible to zinc (Zn) deficiency as a result of their direct
[...] Read more.
A variety of agrochemicals, especially fertilizers, are applied indiscriminately by farmers across trapezoidal landscapes to increase productivity and satisfy the rising food demand. Around one-third of the populace in developing nations is susceptible to zinc (Zn) deficiency as a result of their direct reliance on cereals as a source of calories. Zinc, an essential micronutrient for plants, performs several critical functions throughout the life cycle of a plant. Zinc is frequently disregarded, due to its indirect contribution to the enhancement of yield. Soil Zn deficiency is one of the most prevalent micronutrient deficiencies that reduces crop yield. A deficiency of Zn in both plants and soils results from the presence of Zn in fixed forms that are inaccessible to plants, which characterizes the majority of agricultural soils. As a result, alternative and environmentally sustainable methods are required to satisfy the demand for food. It appears that the application of zinc-solubilizing bacteria (ZSB) for sustainable agriculture is feasible. Inoculating plants with ZSB is likely a more efficacious strategy for augmenting Zn translocation in diverse edible plant components. ZSB possessing plant growth-promoting characteristics can serve as bio-elicitors to promote sustainable plant growth, through various methods that are vital to the health and productivity of plants. This review provides an analysis of the efficacy of ZSB, the functional characteristics of ZSB-mediated Zn localization, the mechanism underlying Zn solubilization, and the implementation of ZSB to increase crop yield.
Full article
(This article belongs to the Special Issue Interaction between Plants and Growth-Promoting Rhizobacteria (PGPR) for Sustainable Development)
►▼
Show Figures
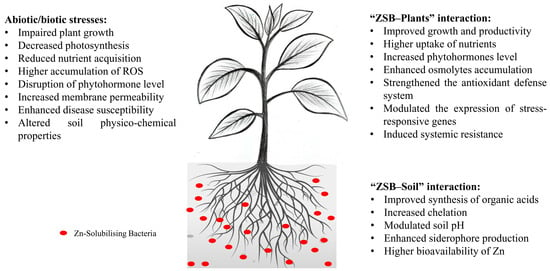
Figure 1
Open AccessArticle
Variations in Morpho-Cultural Characteristics and Pathogenicity of Fusarium moniliforme of Bakanae Disease of Rice and Evaluation of In Vitro Growth Suppression Potential of Some Bioagents
by
Abdullah Al Amin, Md. Hosen Ali, Md. Morshedul Islam, Shila Chakraborty, Muhammad Humayun Kabir and Md. Atiqur Rahman Khokon
Bacteria 2024, 3(1), 1-14; https://doi.org/10.3390/bacteria3010001 - 29 Jan 2024
Abstract
Bakanae is one of the important diseases of rice in Bangladesh that causes substantial yield loss every year. We collected thirty isolates of Fusarium spp. from bakanae-infected rice plants from different agroecological zones of Bangladesh and investigated the variations in cultural and morphological
[...] Read more.
Bakanae is one of the important diseases of rice in Bangladesh that causes substantial yield loss every year. We collected thirty isolates of Fusarium spp. from bakanae-infected rice plants from different agroecological zones of Bangladesh and investigated the variations in cultural and morphological characteristics and pathogenicity. Diversity was found in cultural characteristics, viz., colony features, phialide, chlamydospore formation, shape, and size of macro- and microconidia. Three variants of Fusarium species such as F. moniliforme, F. fujikuroi, and F. proliferatum were identified on PDA media based on their cultural and morphological characteristics. Isolate FM10 (F. moniliforme) exhibited the highest disease aggressiveness in developing elongated plants (26.50 cm), the highest number of chlorotic leaves (5.75), and a lower germination percentage. We evaluated different bioagents against the virulent isolate of F. moniliforme to develop a rice bakanae disease management approach. Four bioagents, viz., Trichoderma spp., Bacillus subtilis, Pseudomonas fluorescens, and Achromobacter spp., were evaluated for growth suppression of F. moniliforme. Among the bioagents, Achromobacter spp. and B. subtilis (BS21) showed 73.54% and 71.61% growth suppression, respectively. The investigation revealed that the application of Achromobacter spp. and B. subtilis (BS21) would be a potential candidate for effective and eco-friendly management of the bakanae disease of rice.
Full article
(This article belongs to the Special Issue Interaction between Plants and Growth-Promoting Rhizobacteria (PGPR) for Sustainable Development)
►▼
Show Figures
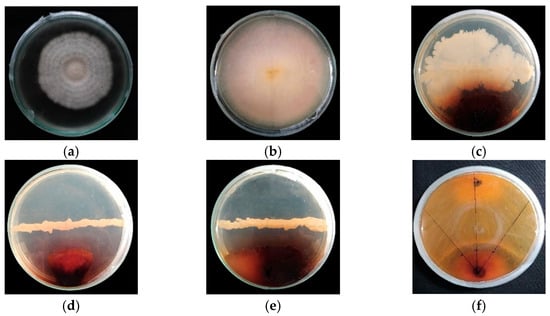
Figure 1
Open AccessCase Report
Bacteremia Following Alkalihalobacillus clausii (Formerly Bacillus clausii) Administration in Immunosuppressed Adults: A Case Series
by
José Pablo Díaz-Madriz, Esteban Zavaleta-Monestel, Carolina Rojas-Chinchilla, Sebastián Arguedas-Chacón, Bruno Serrano-Arias, Mery Alejandra Ferreto-Meza, Betzy María Romero-Chavarría, Priscila Zumbado-Amerling, Ana Fernanda Vásquez-Mendoza, Karla Sofia Gutiérrez-González and César Rodríguez
Bacteria 2023, 2(4), 185-195; https://doi.org/10.3390/bacteria2040014 - 12 Dec 2023
Abstract
(1) Background: Given the widespread use of Alkalihalobacillus clausii (A. clausii) as a probiotic in recent decades and the detection of bacteremia cases in a group of patients, we sought to analyze cases of A. clausii bacteremia following oral probiotic use
[...] Read more.
(1) Background: Given the widespread use of Alkalihalobacillus clausii (A. clausii) as a probiotic in recent decades and the detection of bacteremia cases in a group of patients, we sought to analyze cases of A. clausii bacteremia following oral probiotic use (2) Methods: A retrospective observational study was conducted at a private hospital in San Jose, Costa Rica. Cases of bacteremia caused by A. clausii confirmed by the microbiology laboratory were analyzed in patients who received oral treatment with this probiotic between January 2020 and January 2022. In addition, an isolate (HCB-AC2) was compared through whole genome sequencing to demonstrate the correlation of bacteremia and A. clausii. Possible vulnerability factors related to the development of this condition were determined. (3) Results: Four cases were identified in this hospital over 2 years. Genomic analysis of isolate HCB-AC2, using two different methods, showed identical results. This indicates that HCB-AC2 is genomically identical to ENTpro and the Enterogermina® reference genome. The median age was 71 years, and all patients had some degree of immunosuppression. All patients met at least three sepsis criteria at the time of bacterial identification. Most patients were treated with vancomycin and levofloxacin. Three of the identified patients died. (4) Conclusion: A. clausii can be used as a probiotic, but caution is advised when used in immunosuppressed and elderly patients. These findings align with those reported in similar case studies.
Full article
Open AccessArticle
The Effect of Cell-Free Nontuberculous Mycobacterium Supernatants on Antibiotic Resistance and Biofilm Formation of Opportunistic Pathogens
by
Elena A. Shchuplova and Olga A. Gogoleva
Bacteria 2023, 2(4), 174-184; https://doi.org/10.3390/bacteria2040013 - 01 Dec 2023
Abstract
►▼
Show Figures
The presence of nontuberculous mycobacteria in biofilms on the surface of medical devices may affect the opportunistic pathogens that are common inhabitants of such biofilms. This study assessed the effect of Mycolicibacterium iranicum cell-free supernatants on biofilm formation and antibiotic susceptibility of Escherichia
[...] Read more.
The presence of nontuberculous mycobacteria in biofilms on the surface of medical devices may affect the opportunistic pathogens that are common inhabitants of such biofilms. This study assessed the effect of Mycolicibacterium iranicum cell-free supernatants on biofilm formation and antibiotic susceptibility of Escherichia coli and Staphylococcus epidermidis differing in the anti-hemoglobin activity level. The cell-free supernatants have been shown to stimulate biofilm formation and also help reduce susceptibility of opportunistic pathogens to a number of antibiotics.
Full article
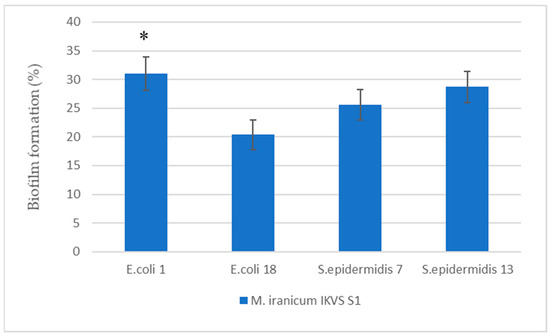
Figure 1
Open AccessArticle
Comprehensive Analysis of Klebsiella pneumoniae Culture Identification and Antibiogram: Implications for Antimicrobial Susceptibility Patterns from Sputum Samples in Mardan, Khyber Pakhtunkhwa, Pakistan
by
Ruby Khan, Saima Wali and Bakht Pari
Bacteria 2023, 2(4), 155-173; https://doi.org/10.3390/bacteria2040012 - 15 Nov 2023
Cited by 1
Abstract
►▼
Show Figures
Klebsiella pneumoniae, a Gram-negative pathogen, poses a significant threat as a cause of community- and hospital-acquired infections worldwide. The emergence of multidrug-resistant strains, particularly in nosocomial settings, has further complicated the management of these infections. This study aimed to investigate the culture
[...] Read more.
Klebsiella pneumoniae, a Gram-negative pathogen, poses a significant threat as a cause of community- and hospital-acquired infections worldwide. The emergence of multidrug-resistant strains, particularly in nosocomial settings, has further complicated the management of these infections. This study aimed to investigate the culture identification and antibiogram of K. pneumoniae isolated from sputum samples collected in various districts in Mardan, Khyber Pakhtunkhwa, Pakistan. A total of 16 sputum samples were collected from patients at the Mardan Medical Complex. Standard microbiological techniques were employed to identify K. pneumoniae, and the antibiotic susceptibility testing was performed using the Kirby–Bauer disc diffusion method, following CLSI guidelines. Among the confirmed K. pneumoniae isolates, approximately 50% were found to be multidrug-resistant. The results indicated resistance to several antibiotics, including vancomycin (30 g), amikacin (30 g), chloramphenicol (30 g), amoxicillin (30 g), and ticarcillin (75 g), while being susceptible to meropenem (10 g), piperacillin (100 g), and tazobactam (110 g). A bioinformatics analysis was also conducted to gain deeper insights into the resistance patterns and potential clustering of isolates. This comprehensive study provides valuable information on the epidemiological trends and antimicrobial susceptibility profile of K. pneumoniae in the region. The findings of this study highlight the urgent need for antimicrobial stewardship programs to combat the rising challenge of antibiotic resistance. Understanding the resistance landscape of K. pneumoniae can guide healthcare professionals in selecting appropriate antibiotics and improving patient outcomes. These data can contribute to the formulation of local antibiotic policies and assist clinicians in making rational choices for antibiotic therapy.
Full article
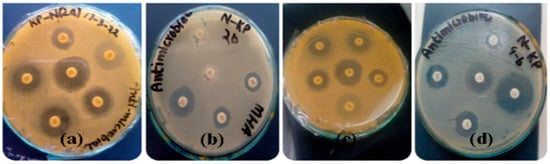
Figure 1
Open AccessArticle
Role of Curing Agents in the Adaptive Response of the Bioprotective Latilactobacillus curvatus CRL 705 from a Physiologic and Proteomic Perspective
by
Lucrecia C. Terán, Alejandra Orihuel, Emilse Bentencourt, Raúl Raya and Silvina Fadda
Bacteria 2023, 2(4), 142-154; https://doi.org/10.3390/bacteria2040011 - 02 Nov 2023
Abstract
►▼
Show Figures
During meat processing, lactic acid bacteria (LAB) have to competitively adapt to the hostile environment produced by curing additives (CA). The objective of this study was to investigate the ability of Latilactobacillus curvatus CRL 705, a bioprotective strain of meat origin, to adapt
[...] Read more.
During meat processing, lactic acid bacteria (LAB) have to competitively adapt to the hostile environment produced by curing additives (CA). The objective of this study was to investigate the ability of Latilactobacillus curvatus CRL 705, a bioprotective strain of meat origin, to adapt to CA. A physiological and proteomic approach was performed. CRL 705 was grown in a chemically defined medium (CDM) containing specific concentrations of CA (NaCl, nitrite, sucrose, and ascorbic acid). The results showed minor differences in growth kinetics in the presence of CA. Glucose consumption, present in the CDM, and production of lactic acid and bacteriocins were not significantly affected. Proteomic analyses indicated that most of the identified proteins (36 out of 39) mainly related to carbohydrate metabolism (18%), posttranslational modifications (15.6%), energy production and conversion (11.1%), translation (11.1%), and nucleotide metabolism (8.9%) were underexpressed. In response to the studied CA, CRL 705 slowed down its general metabolism, achieving slight changes in physiological and proteomic parameters. The observed performance is another characteristic that extends the well-known competitive profile of CRL 705 as a meat starter and bioprotective culture. This is the first report dealing with the impact of CA on LAB proteomics.
Full article
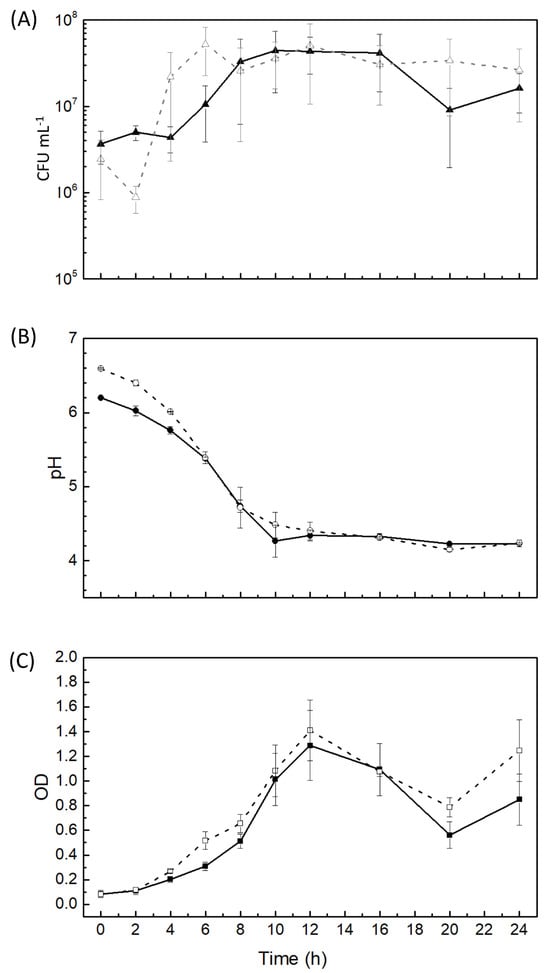
Figure 1
Open AccessCommunication
Enhancing Manganese Availability for Plants through Microbial Potential: A Sustainable Approach for Improving Soil Health and Food Security
by
Bahman Khoshru, Debasis Mitra, Alireza Fallah Nosratabad, Adel Reyhanitabar, Labani Mandal, Beatrice Farda, Rihab Djebaili, Marika Pellegrini, Beatriz Elena Guerra-Sierra, Ansuman Senapati, Periyasamy Panneerselvam and Pradeep Kumar Das Mohapatra
Bacteria 2023, 2(3), 129-141; https://doi.org/10.3390/bacteria2030010 - 06 Aug 2023
Cited by 2
Abstract
Manganese (Mn) is essential for plant growth, as it serves as a cofactor for enzymes involved in photosynthesis, antioxidant synthesis, and defense against pathogens. It also plays a role in nutrient uptake, root growth, and soil microbial communities. However, the availability of Mn
[...] Read more.
Manganese (Mn) is essential for plant growth, as it serves as a cofactor for enzymes involved in photosynthesis, antioxidant synthesis, and defense against pathogens. It also plays a role in nutrient uptake, root growth, and soil microbial communities. However, the availability of Mn in the soil can be limited due to factors like soil pH, redox potential, organic matter content, and mineralogy. The excessive use of chemical fertilizers containing Mn can lead to negative consequences for soil and environmental health, such as soil and water pollution. Recent research highlights the significance of microbial interactions in enhancing Mn uptake in plants, offering a more environmentally friendly approach to address Mn deficiencies. Microbes employ various strategies, including pH reduction, organic acid production, and the promotion of root growth, to increase Mn bioavailability. They also produce siderophores, anti-pathogenic compounds, and form symbiotic relationships with plants, thereby facilitating Mn uptake, transport, and stimulating plant growth, while minimizing negative environmental impacts. This review explores the factors impacting the mobility of Mn in soil and plants, and highlights the problems caused by the scarcity of Mn in the soil and the use of chemical fertilizers, including the consequences. Furthermore, it investigates the potential of different soil microbes in addressing these challenges using environmentally friendly methods. This review suggests that microbial interactions could be a promising strategy for improving Mn uptake in plants, resulting in enhanced agricultural productivity and environmental sustainability. However, further research is needed to fully understand these interactions’ mechanisms and optimize their use in agricultural practices.
Full article
(This article belongs to the Special Issue Interaction between Plants and Growth-Promoting Rhizobacteria (PGPR) for Sustainable Development)
►▼
Show Figures
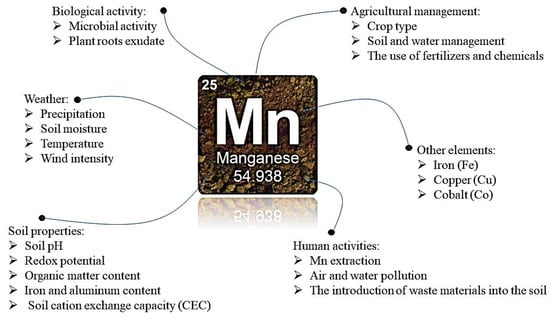
Figure 1
Open AccessArticle
Geochemical and Microbiological Composition of Soils and Tailings Surrounding the Komsomolsk Tailings, Kemerovo Region, Russia
by
Natalya Abrosimova, Svetlana Bortnikova, Alexey Edelev, Valery Chernukhin, Alexander Reutsky, Nikolay Abrosimov and Ivan Gundyrev
Bacteria 2023, 2(3), 116-128; https://doi.org/10.3390/bacteria2030009 - 09 Jul 2023
Cited by 1
Abstract
►▼
Show Figures
Microorganisms have the potential to address environmental pollution, but the interaction mechanism between microorganisms and mine tailings is not well understood. This work was aimed at determining the bacterial isolates in soils and mine tailings and evaluating the distribution of metals, antimony (Sb),
[...] Read more.
Microorganisms have the potential to address environmental pollution, but the interaction mechanism between microorganisms and mine tailings is not well understood. This work was aimed at determining the bacterial isolates in soils and mine tailings and evaluating the distribution of metals, antimony (Sb), and arsenic (As) in the soils around the Komsomolsk tailings. Areas with high concentrations of As, Sb, cadmium (Cd), and lead (Pb) were found. Assessment based on the value of the contamination factor (CF) indicated large-scale As, Sb, Pb, Cd, iron (Fe), bismuth (Bi), and beryllium (Be) pollution, especially in soils sampled from the northeast direction of the mine tailings. Soils had a higher number of CFUs per g of dry weight than did the tailings, ranging from 84 × 106 to 3.1 × 109 and from 20 × 106 to 1.7 × 109, respectively. Arsenic exhibited a positive statistical correlation with the number of CFUs of Agrococcus and Staphylococcus. In addition, a positive correlation was found between the concentration of Co and the number of CFUs of Moraxella and Microbacterium. The Sb exhibited a positive correlation with Streptomyces. These results can be used to develop methods for waste reclamation, including the use of isolated bacterial strains for arsenic removal by precipitation.
Full article
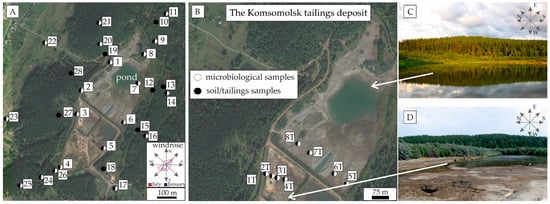
Figure 1
Open AccessReview
Rock Phosphate Solubilizing Potential of Soil Microorganisms: Advances in Sustainable Crop Production
by
Bahman Khoshru, Alireza Fallah Nosratabad, Debasis Mitra, Manju Chaithra, Younes Rezaee Danesh, Gökhan Boyno, Sourav Chattaraj, Ankita Priyadarshini, Snežana Anđelković, Marika Pellegrini, Beatriz Elena Guerra-Sierra and Somya Sinha
Bacteria 2023, 2(2), 98-115; https://doi.org/10.3390/bacteria2020008 - 10 May 2023
Cited by 3
Abstract
Phosphorus (P) is one of the most important elements required for crop production. The ideal soil pH for its absorption by plants is about 6.5, but in alkaline and acidic soils, most of the consumed P forms an insoluble complex with calcium, iron,
[...] Read more.
Phosphorus (P) is one of the most important elements required for crop production. The ideal soil pH for its absorption by plants is about 6.5, but in alkaline and acidic soils, most of the consumed P forms an insoluble complex with calcium, iron, and aluminum elements and its availability for absorption by the plant decreases. The supply of P needed by plants is mainly achieved through chemical fertilizers; however, in addition to the high price of these fertilizers, in the long run, their destructive effects will affect the soil and the environment. The use of cheap and abundant resources such as rock phosphate (RP) can be an alternative strategy for P chemical fertilizers, but the solubilization of P of this source has been a challenge for agricultural researchers. For this, physical and chemical treatments have been used, but the solution that has recently attracted the attention of the researchers is to use the potential of rhizobacteria to solubilize RP and supply P to plants by this method. These microorganisms, via. mechanisms such as proton secretion, organic and mineral acid production, siderophore production, etc., lead to the solubilization of RP, and by releasing its P, they improve the quantitative and qualitative performance of agricultural products. In this review, addressing the potential of rhizosphere microbes (with a focus on rhizobacteria) as an eco-friendly strategy for RP solubilization, along with physical and chemical solutions, has been attempted.
Full article
(This article belongs to the Special Issue Interaction between Plants and Growth-Promoting Rhizobacteria (PGPR) for Sustainable Development)
►▼
Show Figures
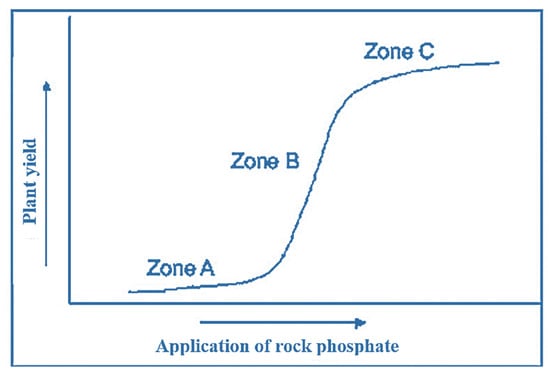
Figure 1
Open AccessArticle
The Isolation, Screening, and Characterization of Polyhydroxyalkanoate-Producing Bacteria from Hypersaline Lakes in Kenya
by
Martin N. Muigano, Sylvester E. Anami, Justus M. Onguso and Godfrey M. Omare
Bacteria 2023, 2(2), 81-97; https://doi.org/10.3390/bacteria2020007 - 08 May 2023
Cited by 1
Abstract
►▼
Show Figures
Extremophilic microorganisms such as those that thrive in high-salt and high-alkaline environments are promising candidates for the recovery of useful biomaterials including polyhydroxyalkanoates (PHAs). PHAs are ideal alternatives to synthetic plastics because they are biodegradable, biocompatible, and environmentally friendly. This work was aimed
[...] Read more.
Extremophilic microorganisms such as those that thrive in high-salt and high-alkaline environments are promising candidates for the recovery of useful biomaterials including polyhydroxyalkanoates (PHAs). PHAs are ideal alternatives to synthetic plastics because they are biodegradable, biocompatible, and environmentally friendly. This work was aimed at conducting a bioprospection of bacteria isolated from hypersaline-alkaliphilic lakes in Kenya for the potential production of PHAs. In the present study, 218 isolates were screened by Sudan Black B and Nile Red A staining. Of these isolates, 31 were positive for PHA production and were characterized using morphological, biochemical, and molecular methods. Through 16S rRNA sequencing, we found that the isolates belonged to the genera Arthrobacter spp., Bacillus spp., Exiguobacterium spp., Halomonas spp., Paracoccus spp., and Rhodobaca spp. Preliminary experiments revealed that Bacillus sp. JSM-1684023 isolated from Lake Magadi had the highest PHA accumulation ability, with an initial biomass-to-PHA conversion rate of 19.14% on a 2% glucose substrate. Under optimized fermentation conditions, MO22 had a maximum PHA concentration of 0.516 g/L from 1.99 g/L of cell dry weight and 25.9% PHA conversion, equivalent to a PHA yield of 0.02 g/g of biomass. The optimal PHA production media had an initial pH of 9.0, temperature of 35 °C, salinity of 3%, and an incubation period of 48 h with 2.5% sucrose and 0.1% peptone as carbon and nitrogen sources, respectively. This study suggests that bacteria isolated from hypersaline and alkaliphilic tropical lakes are promising candidates for the production of polyhydroxyalkanoates.
Full article

Figure 1
Open AccessCommunication
Enumerating Indigenous Arbuscular Mycorrhizal Fungi (AMF) Associated with Three Permanent Preservation Plots of Tropical Forests in Bangalore, Karnataka, India
by
Saritha Boya, Poorvashree Puttaswamy, Nethravathi Mahadevappa, Balasubramanya Sharma and Remadevi Othumbamkat
Bacteria 2023, 2(1), 70-80; https://doi.org/10.3390/bacteria2010006 - 13 Mar 2023
Cited by 1
Abstract
The establishment of Permanent Preservation Plots (PPPs) in natural forests has a signifi-cant role in assessing the impact of climate change on forests. To pursue long-term studies on cli-mate change, PPPs were established during the year 2016 in two major forest areas in
[...] Read more.
The establishment of Permanent Preservation Plots (PPPs) in natural forests has a signifi-cant role in assessing the impact of climate change on forests. To pursue long-term studies on cli-mate change, PPPs were established during the year 2016 in two major forest areas in Bangalore to conduct ecological studies to monitor the vegetation changes. One of the objectives of the study was to understand the drivers of diversity, such as soils, in terms of nutrients and physical and biological properties. The native tropical forest of Bangalore, which houses Bannerghatta National Park (BNP) on the outskirts, is relatively underexplored in terms of its microflora, particularly arbuscular my-corrhizal fungi (AMF). Hence, the present study was aimed at the quantitative estimation of arbus-cular mycorrhizal fungi (AMF) in the three 1-ha PPPs which were established in Bannerughatta National Park (BNP) and Doresanipalya Reserve Forest (DRF) as per the Centre for Tropical Forest Sciences (CTFS) protocol. In BNP, two plots were established, one in the Thalewood house area (mixed, moist, deciduous type) and the other in the Bugurikallu area (dry, deciduous type). In DRF, one plot was established in dry, deciduous vegetation. Each one-hectare plot (100 m × 100 m) was subdivided into twenty-five sub-plots (20 m × 20 m). Composite soil samples were collected during two seasons (dry and wet) and analyzed for AMF spore and available phosphorus (P) content. The results revealed the presence of AMF in all the three plots. Doresanipalya plo had the highest spore number, followed by the Bugurikallu plot and Thalewood house plot. The available phosphorous and AMF spore numbers showed correlations in all the three plots. Among the AMF spores, the Glomus species was found to dominate in all the three plots. The study shows that the dry, decidu-ous forests accommodated more AMF spores than the mixed, moist forests.
Full article
(This article belongs to the Special Issue Interaction between Plants and Growth-Promoting Rhizobacteria (PGPR) for Sustainable Development)
►▼
Show Figures
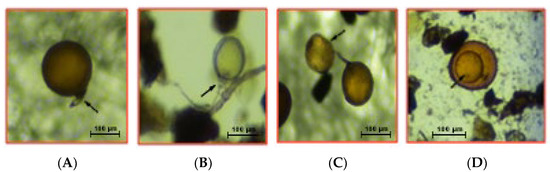
Figure 1
Open AccessArticle
High Fecal Carriage of Extended-Spectrum β-Lactamase Producing Enterobacteriaceae by Children Admitted to the Pediatric University Hospital Complex in Bangui, Central African Republic
by
Hugues Sanke-Waïgana, Cheikh Fall, Jean-Chrysostome Gody, Eliot Kosh Komba, Gilles Ngaya, Jean-Robert Mbecko, Brice Martial Yambiyo, Alexandre Manirakiza, Guy Vernet, Alioune Dieye and Yakhya Dieye
Bacteria 2023, 2(1), 60-69; https://doi.org/10.3390/bacteria2010005 - 01 Mar 2023
Abstract
Antimicrobial resistance (AMR) is a global public health threat. Quality data on AMR are needed to tackle the rise of multidrug-resistant clones. These data are rare in low-income countries, especially in sub-Saharan Africa. In this study, we investigated the rise of extended-spectrum β-lactamase–producing
[...] Read more.
Antimicrobial resistance (AMR) is a global public health threat. Quality data on AMR are needed to tackle the rise of multidrug-resistant clones. These data are rare in low-income countries, especially in sub-Saharan Africa. In this study, we investigated the rise of extended-spectrum β-lactamase–producing (ESBL) Enterobacteriaceae in Bangui, Central African Republic. We collected 278 fecal samples from 0–5-year-old children admitted to the Pediatric University Hospital Complex in Bangui from July to September 2021. Enterobacteriaceae were isolated and identified, and their susceptibility to 19 antibiotics was tested. We recovered one and two Enterobacteriaceae species from 208 and 29 samples, respectively. One clone of each species from each sample was further characterized, for a total of 266 isolates. Escherichia coli predominated, followed by Klebsiella. AMR was frequent, with 98.5% (262/266) of the isolates resistant to at least one antibiotic. Additionally, 89.5% (238/266) of the isolates were multidrug resistant, with resistance being frequent against all tested antibiotics except carbapenems and tigecycline, for which no resistance was found. Importantly, 71.2% (198/278) of the children carried at least one ESBL species, and 85.3% (227/266) of the isolates displayed this phenotype. This study confirms the rise of ESBL Enterobacteriaceae in Bangui and stresses the need for action to preserve the efficacy of antibiotics, as crucial for the treatment of bacterial infections.
Full article
Open AccessArticle
Effect of Lactic Acid Fermentation on Phytochemical Content, Antioxidant Capacity, Sensory Acceptability and Microbial Safety of African Black Nightshade and African Spider Plant Vegetables
by
Marie Lys Irakoze, Eliud Nalianya Wafula and Eddy Elkana Owaga
Bacteria 2023, 2(1), 48-59; https://doi.org/10.3390/bacteria2010004 - 11 Feb 2023
Cited by 2
Abstract
►▼
Show Figures
Traditional preparation of African indigenous vegetables (AIVs) such as African black nightshade (Solanum nigrum) and African spiderplant (Cleome gynandra) involves either boiling and discarding the first water or lengthy boiling. Fermentation is considered a better alternative processing technique due
[...] Read more.
Traditional preparation of African indigenous vegetables (AIVs) such as African black nightshade (Solanum nigrum) and African spiderplant (Cleome gynandra) involves either boiling and discarding the first water or lengthy boiling. Fermentation is considered a better alternative processing technique due to the enhanced retention of phytochemical contents and sensory properties. However, little is known about the impact of lactic acid fermentation on the phytochemical content, antioxidant capacity, sensory acceptability and microbial safety of the African black nightshade and African spiderplant. This study aimed to ferment AIVs using combined starter cultures (Lactobacillus fermentum and Lactococcus lactis) and further determine their effect on the phytochemical content (phenolic compounds and flavonoids), antioxidant capacity, sensory acceptability and microbial safety of the vegetables. There was a marked increase in phenol and flavonoid contents in all fermented vegetables (p < 0.05). The highest phenol content was 228.8 mg/g GAE (gallic acid equivalent) in the starter-culture-inoculated African black nightshade, while flavonoid content was 10.6 mg/g QE (quercetin equivalent) in the same. Starter-culture-inoculated AIVs presented significantly higher antioxidant capacity with a 60–80% radical scavenging activity compared to levels in uninoculated batches (p < 0.05). Fermented vegetables were more liked than the boiled vegetables and were microbiologically safe. In conclusion, lactic fermentation of AIVs increased phytochemical contents (phenolic compounds and flavonoids), maintained antioxidant capacity and improved product safety and sensory acceptability. Therefore, fermentation and consumption of the African indigenous vegetables are to be encouraged.
Full article
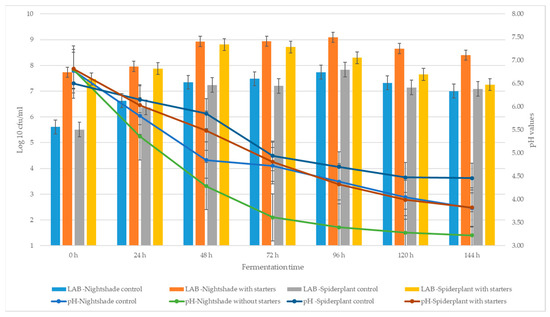
Figure 1
Open AccessArticle
Intestinal Carriage of Extended Spectrum Beta-Lactamase-Producing Salmonella enterica from Chickens and Poultry Farmers in Dschang, in the Western Region of Cameroon
by
Cecile Ingrid Djuikoue, Cedric Dylan Seugnou Nana, Joelle Nzenya, Charlene Tomi, Noemy Chounna, Olivier Pomte, Benjamin D. Thumamo Pokam and Teke Apalata
Bacteria 2023, 2(1), 37-47; https://doi.org/10.3390/bacteria2010003 - 20 Jan 2023
Abstract
►▼
Show Figures
Salmonella enterica is the principal causative agent of salmonellosis, a threat to human health. Because of its high antimicrobial resistance potential, Salmonella enterica has become worrisome, mostly in developing countries where hygiene and antimicrobial usage are defective. This study aimed to determine the
[...] Read more.
Salmonella enterica is the principal causative agent of salmonellosis, a threat to human health. Because of its high antimicrobial resistance potential, Salmonella enterica has become worrisome, mostly in developing countries where hygiene and antimicrobial usage are defective. This study aimed to determine the epidemiology of the intestinal carriage of Extended Spectrum β-Lactamase producing Salmonella enterica from chickens and poultry farmers in Dschang, a town in the western region of Cameroon. A total of 416 chickens and 72 farmers were sampled between May and October 2020; and Salmonella enterica were isolated and subjected to extended spectrum β-lactamase screening. Logistic regression was used to test for statistical associations using a p-value of ≤0.05. Results from this study revealed that the prevalence of the intestinal carriage of Salmonella enterica for chickens and farmers were 55.77% [51.00; 60.54] and 22.22% [12.62; 31.82], respectively. Meanwhile, the intestinal carriage of Extended Spectrum β-Lactamase producing Salmonella enterica was 23.08% [13.76; 32.40] and 5.55% [0.26; 10.84] from chickens and poultry farmers, respectively. The risk factor for this carriage was revealed to be lack of knowledge by actors in livestock industries of antibiotic resistance. Chickens, just like poultry farmers, represent the starting point of community salmonellosis, which is difficult to cure; therefore, sensitization of breeders is an effective tool for the mitigation of this burden.
Full article
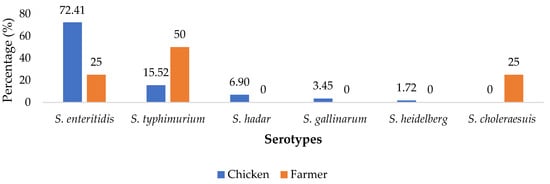
Figure 1
Open AccessReview
Role and Regulation of Clp Proteases: A Target against Gram-Positive Bacteria
by
Camila Queraltó, Ricardo Álvarez, Constanza Ortega, Fernando Díaz-Yáñez, Daniel Paredes-Sabja and Fernando Gil
Bacteria 2023, 2(1), 21-36; https://doi.org/10.3390/bacteria2010002 - 06 Jan 2023
Cited by 3
Abstract
►▼
Show Figures
Bacterial proteases participate in the proteolytic elimination of misfolded or aggregated proteins, carried out by members of the AAA+ protein superfamily such as Hsp100/Clp, Lon, and FtsH. It is estimated that the Clp and Lon families perform around 80% of cellular proteolysis in
[...] Read more.
Bacterial proteases participate in the proteolytic elimination of misfolded or aggregated proteins, carried out by members of the AAA+ protein superfamily such as Hsp100/Clp, Lon, and FtsH. It is estimated that the Clp and Lon families perform around 80% of cellular proteolysis in bacteria. These functions are regulated, in part, through the spatial and/or temporal use of adapter proteins, which participate in the recognition and delivery of specific substrate proteins to proteases. The proteolysis plays an important role in maintaining and controlling the quality of the proteins, avoiding the accumulation and aggregation of unfolded or truncated proteins. However, this is not their only function, since they play an important role in the formation of virulent phenotypes and in the response to different types of stress faced when entering the host or that occur in the environment. This review summarizes the structural and functional aspects of the Clp proteases and their role in Gram-positive microorganisms.
Full article

Figure 1
Open AccessArticle
Isolation and Identification of Autochthonous Lactic Acid Bacteria from Commonly Consumed African Indigenous Leafy Vegetables in Kenya
by
Eliud N. Wafula, Josiah O. Kuja, Tofick B. Wekesa and Paul M. Wanjala
Bacteria 2023, 2(1), 1-20; https://doi.org/10.3390/bacteria2010001 - 05 Jan 2023
Cited by 3
Abstract
►▼
Show Figures
African indigenous leafy vegetables (AILVs) are plants that have been part of the food systems in Sub-Saharan Africa (SSA) for a long time and their leaves, young shoots, flowers, fruits and seeds, stems, tubers, and roots are consumed. These vegetables are high in
[...] Read more.
African indigenous leafy vegetables (AILVs) are plants that have been part of the food systems in Sub-Saharan Africa (SSA) for a long time and their leaves, young shoots, flowers, fruits and seeds, stems, tubers, and roots are consumed. These vegetables are high in vitamins, minerals, protein, and secondary metabolites that promote health. This study aimed at isolating, characterizing, and identifying dominant lactic acid bacteria (LAB) from naturally fermenting commonly consumed AILV in Kenya. A total of 57 LAB strains were isolated and identified based on phenotypic and 16S rRNA gene analyses from three AILVs (23 nightshade leaves, 19 cowpeas leaves, and 15 vegetable amaranth). The highest microbial counts were recorded between 48 h and 96 h of fermentation in all AILVs ranging from approximately log 8 to log 9 CFU/mL with an average pH of 3.7. Fermentation of AILVs was dominated by twenty eight Lactobacillus spp. [Lactiplantibacillus plantarum (22), Limosilactobacillus fermentum (3), Lactiplantibacillus pentosus (2) and Lactiplantibacillus casei (1)], eleven Weissella spp. (Weissella cibaria (8), W. confusa (2), and W. muntiaci) six Leuconostoc spp. [Leuconostoc mesenteroides (3), Leuc. citreum (2) and Leuc. lactis (1)], six Pediococcus pentosaceus, four Enterococcus spp. [Enterococcus mundtii (2), E. faecalis (1) and E. durans (1)] and, finally, two Lactococcus garvieae. These bacteria strains are commonly used in food fermentation as starter cultures and as potential probiotics.
Full article
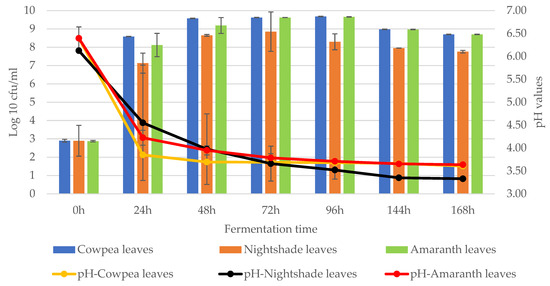
Figure 1
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Biomolecules, Foods, Metabolites, Microorganisms, Pathogens, Bacteria
Bioinformatics, Machine Learning and Risk Assessment in Food Industry
Topic Editors: Bing Niu, Suren Rao Sooranna, Pufeng DuDeadline: 31 July 2025

Conferences
Special Issues
Special Issue in
Bacteria
Interaction between Plants and Growth-Promoting Rhizobacteria (PGPR) for Sustainable Development
Guest Editors: Marika Pellegrini, Beatriz E. Guerra-Sierra, Debasis MitraDeadline: 1 May 2024