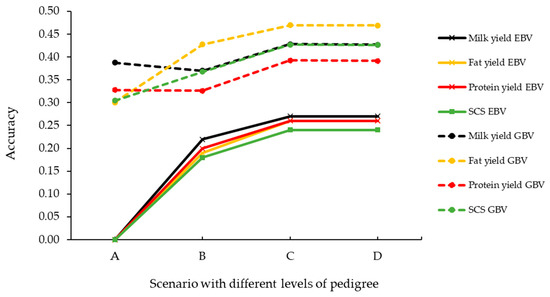
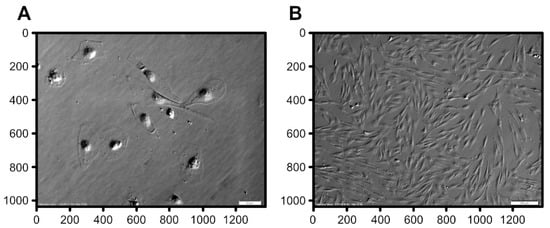
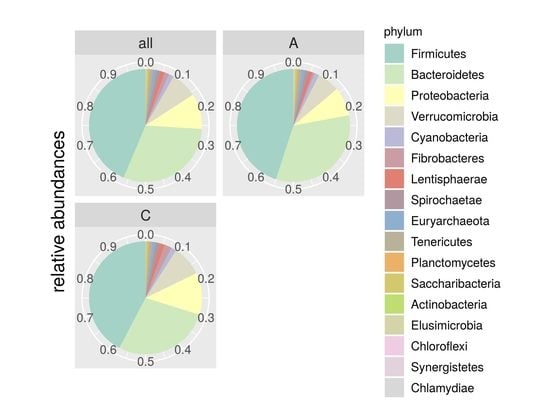
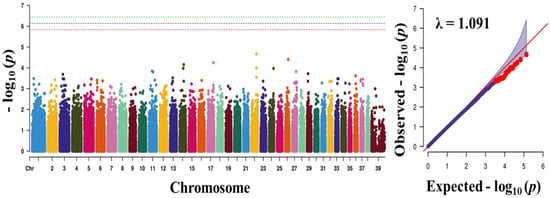
Genomics, Transcriptomics, and Computational Biology for Biodiversity Studies and Quality-Related Traits Selection in Livestock
A topical collection in Animals (ISSN 2076-2615). This collection belongs to the section "Animal Genetics and Genomics".
Viewed by 105615Editor
Topical Collection Information
Dear Colleagues,
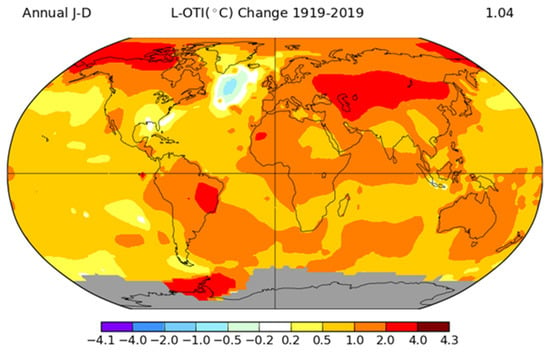
Research efforts in livestock include the identification of the chromosomal regions and candidate genes related to novel complex and economically important traits, together with biodiversity and local breeds preservation.
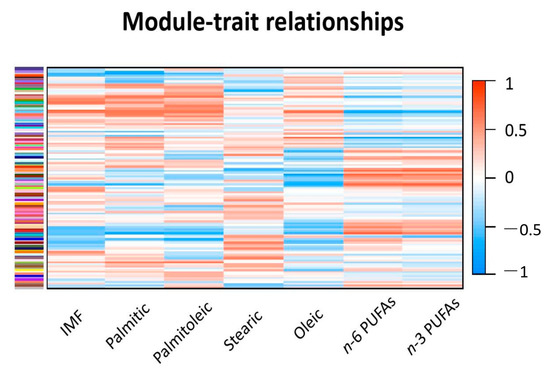
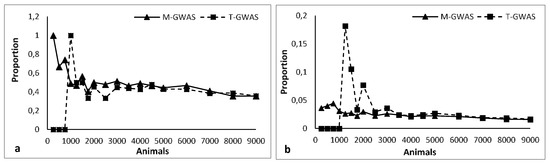
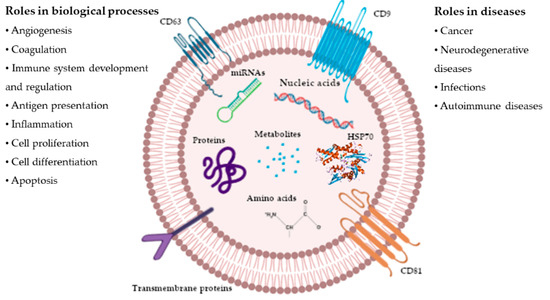
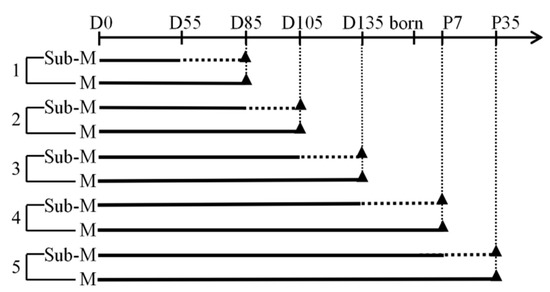
Taking advantage of modern multi-omic approaches for complex traits like high-throughput genotyping, whole-genome sequencing, expression analysis (e.g., GWA studies and RNA sequencing analysis) and so on, it is useful to understand the degree of activation of the metabolic and physiological mechanisms involved with the traits of interest.
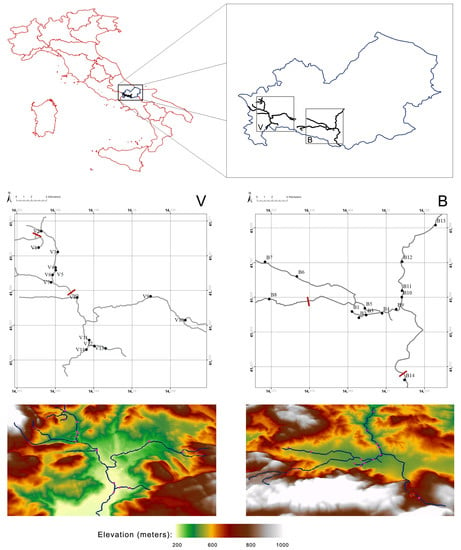
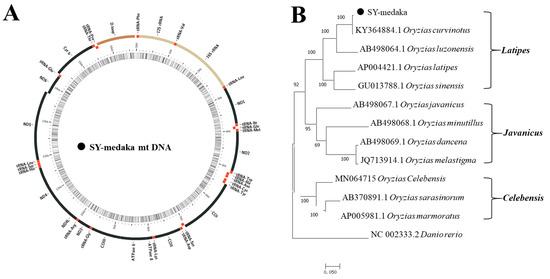
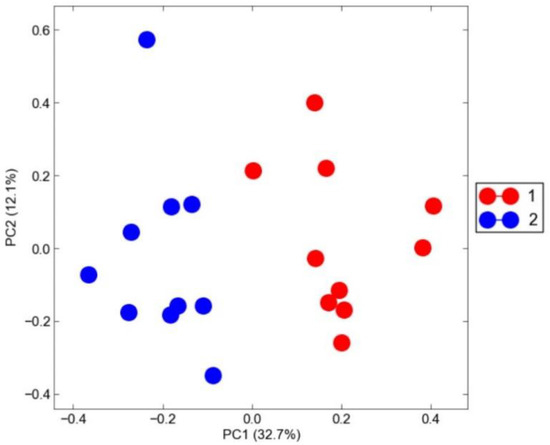
Additionally, molecular data are useful for biodiversity characterization, with the final purpose to gain knowledge about phylogenetic and molecular evolution, and the origin of breeds, so as to assess and exploit the genetic diversity of animal populations, in order to help the development of new selection programs using local breeds.
Moreover, performing studies at a DNA and RNA level, among breeds or populations, involves the use and the development of computational–bioinformatics tools to highlight, interpret, and visualize the results.
We invite original research papers on animal breeding, population genetics, genetic association studies, transcriptomics, and quantitative genomics in livestock, which address the biological mechanisms underlying the expression of complex (quantitative) traits, to give insight into the origin and impact of genetic variation and population stratification at the genome scale, so as to explore genomics opportunities for long-term selection strategies.
Dr. Mariasilvia D'Andrea
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Animals is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2400 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- genetics
- genomics
- transcriptomics
- animal breeding and selection
- population genetics
- novel candidate genes
- GWAS
- RNAseq
- computational biology