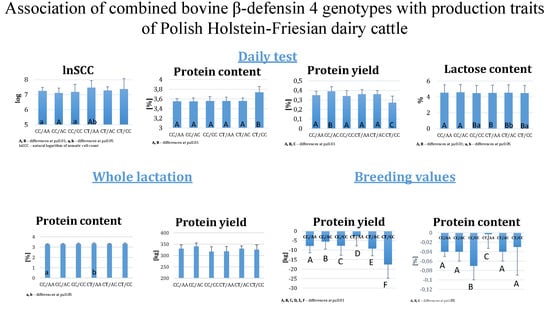
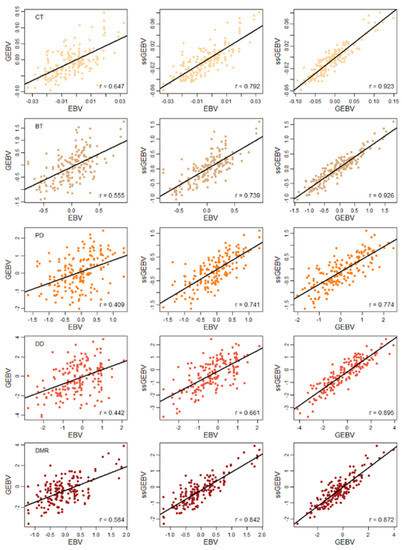
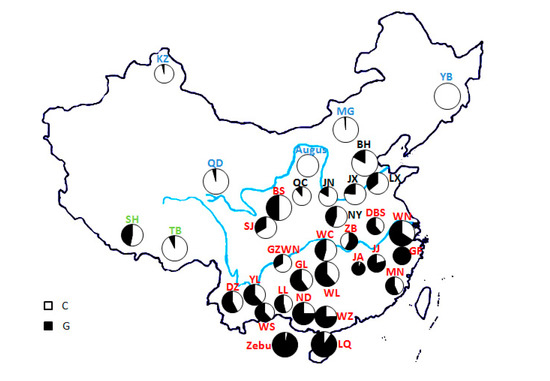
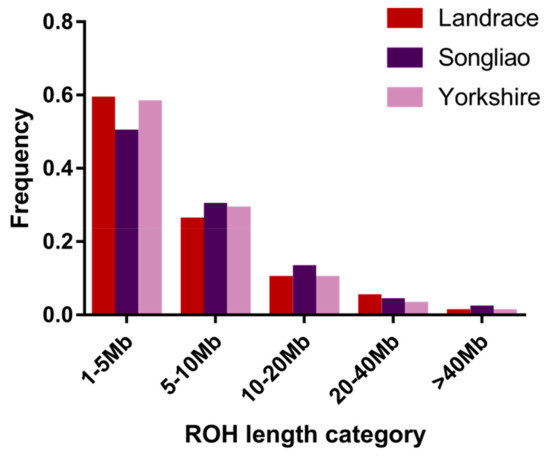
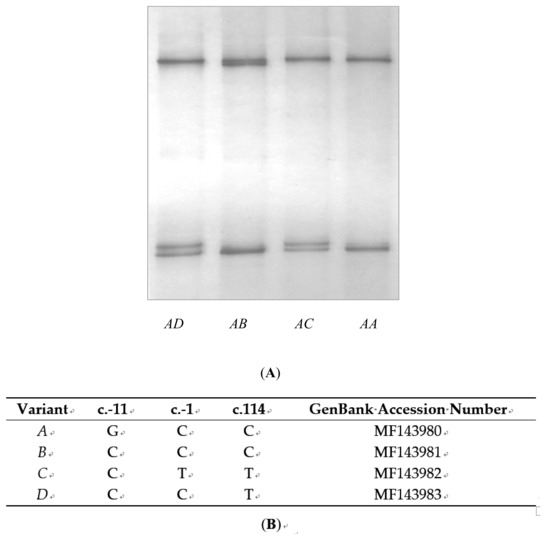
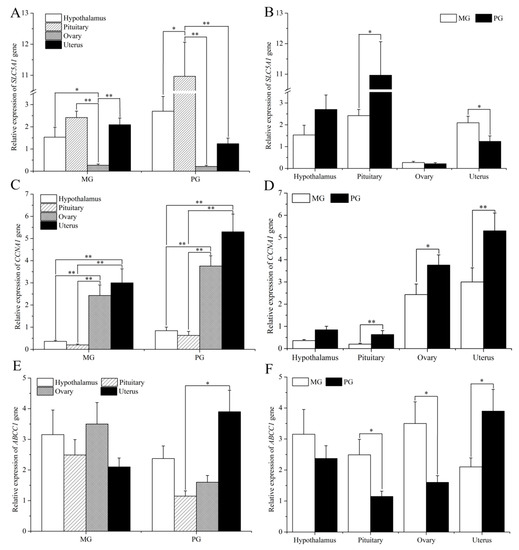
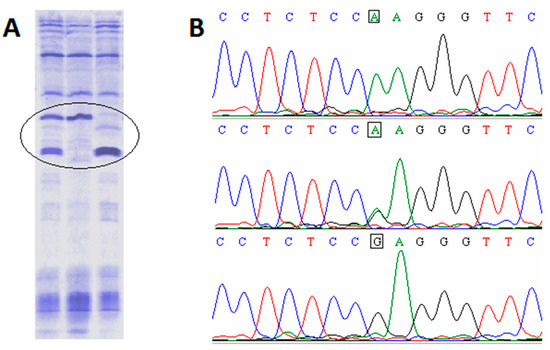
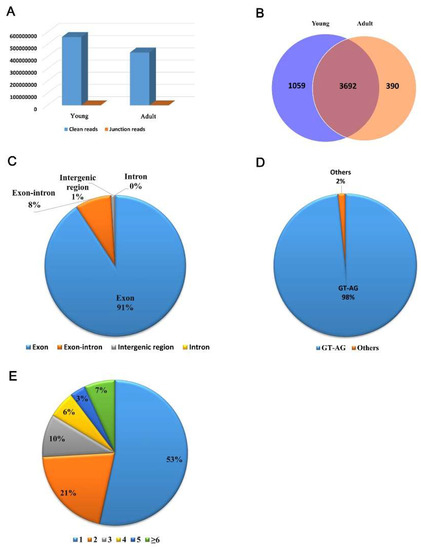
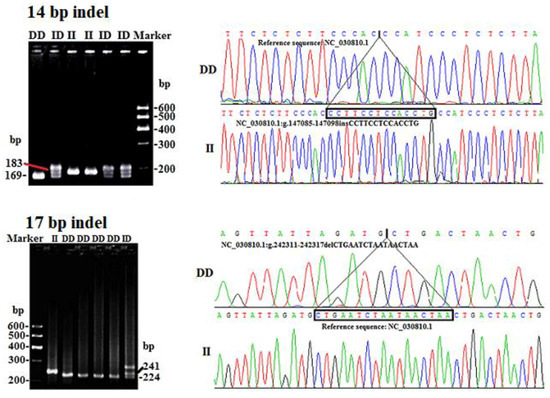
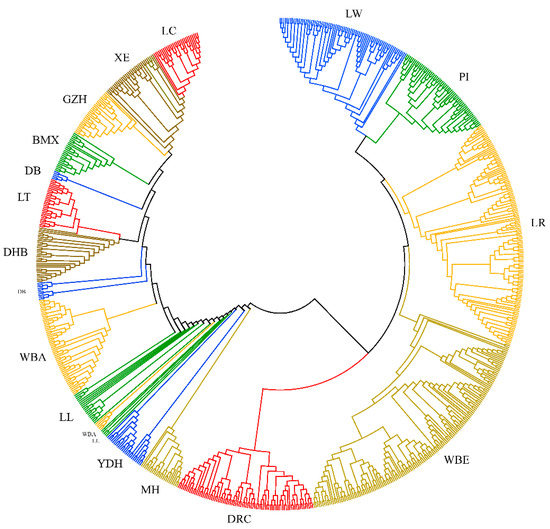
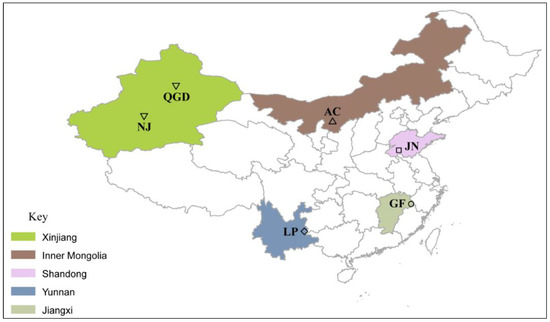
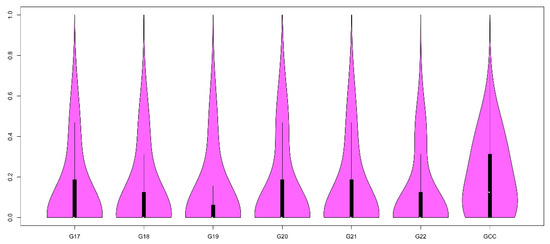
Applications of Quantitative Genetics in Livestock Production
A topical collection in Animals (ISSN 2076-2615). This collection belongs to the section "Animal Genetics and Genomics".
Viewed by 188732Editor
Interests: beef; dairy; genetics; genetic engineering; animal breeding; biostatistics; animal genetics
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
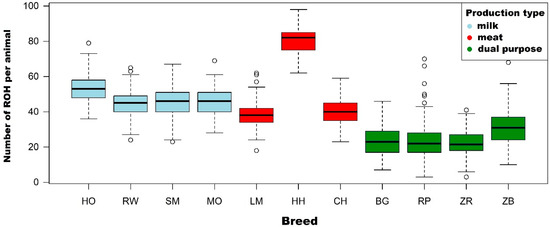
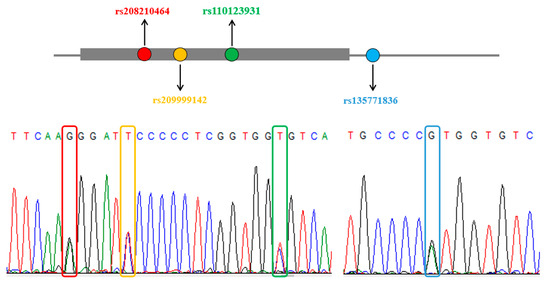
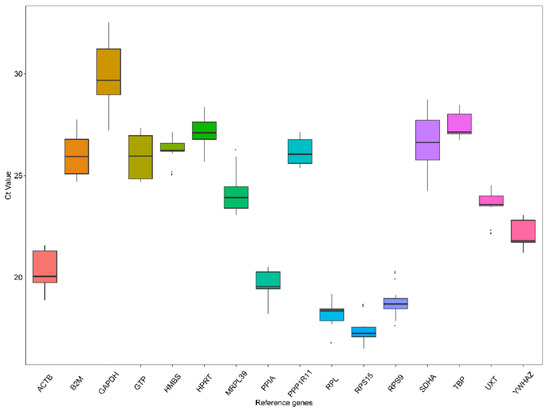
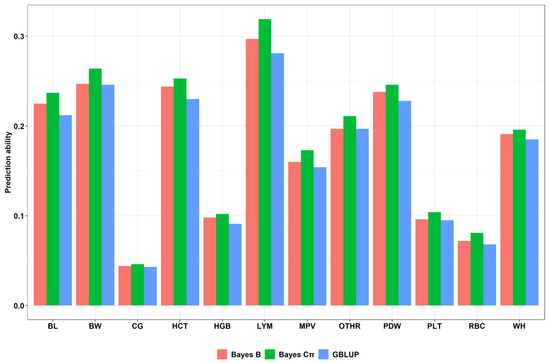
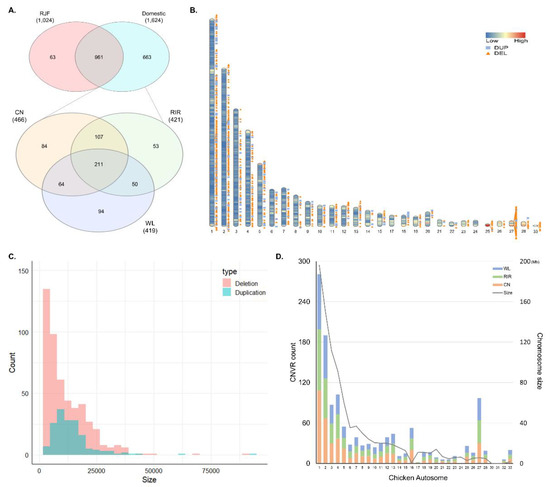
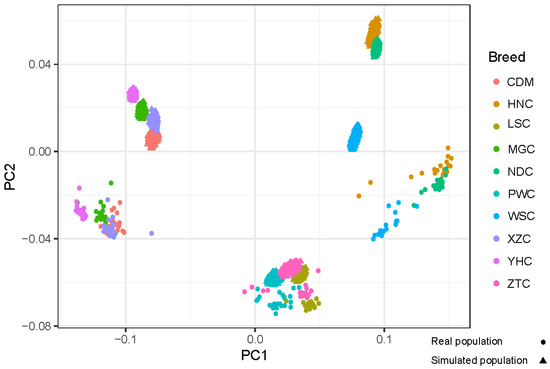
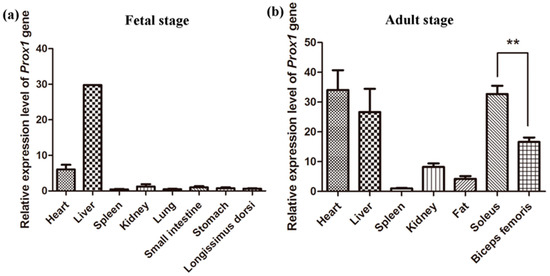
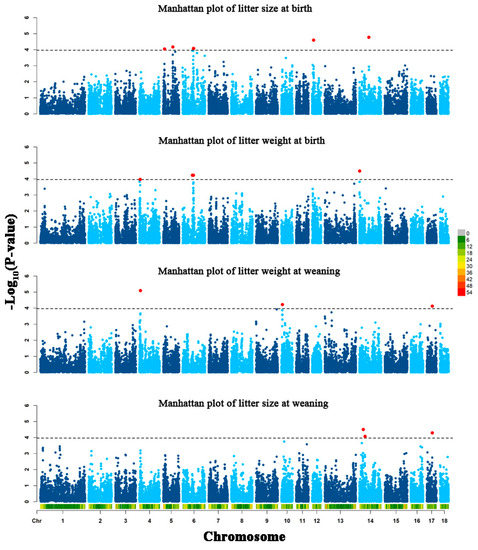
The genetic improvement of livestock in the last 100 years has been remarkable. Most of this genetic change has been due to the application of quantitative genetics. During the 1920s and 1930s, population genetics came of age as the leaders in the field sought to describe genes and chromosomes. Following World War II, J. L. Lush, L. N. Hazel, and others developed the concepts of selection indices and breeding values. In the late 1960s and early 1970s, computing technology advanced to the point where breed associations were able to be generated across herd genetic evaluations using the selection index theory and the mixed model equations first suggested by C. R. Henderson. In the 1980s, BLUP the methodology and reduced animal model were developed, thus allowing for the estimation of expected progeny differences and predicting the transmitting abilities within the breeds of livestock. Another major event in the 1980s was the launch of the genomics era. The genomes of many livestock species have been sequenced, and a large number of evenly spaced genetic markers have been identified. The availability of marker panels of thousands of SNPs, along with new computing technologies, such as a single-step approach to incorporating genomic information into EPDs, have made genomic selection a reality, and have greatly increased the accuracy of selection, even for young animals and for difficult to measure traits such as disease resistance, longevity, and feed efficiency. Much has been accomplished, but much work remains for quantitative geneticists who work with livestock. The commercial genotyping of livestock has been accomplished for millions of animals. Therefore, vast amounts of genomic data remain to be mined. Much remains to be learned concerning the genetic architecture of complex traits and how this knowledge can be applied to livestock production.
Prof. Michael E. Davis
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Animals is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2400 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- population genetics
- animal breeding
- quantitative genetics
- heritability
- genetic variation
- gene frequency
- selection
- mating systems
- genetic improvement
- breeding value.