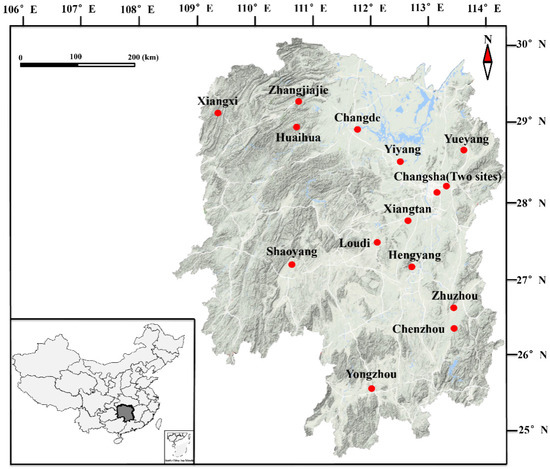
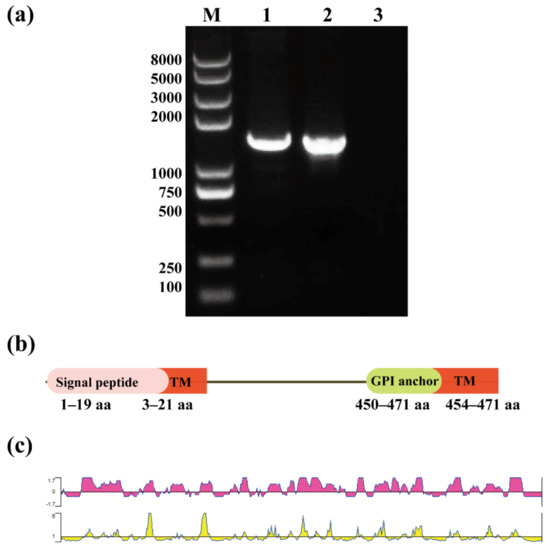
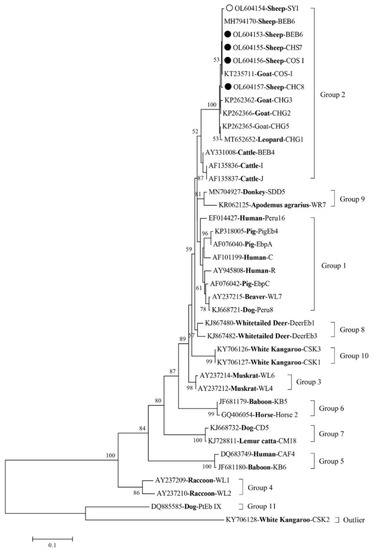
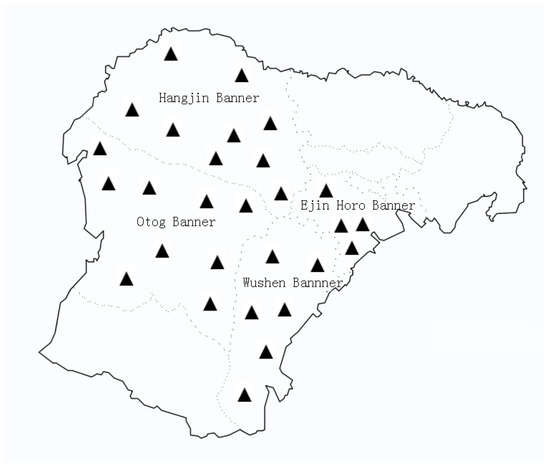
Advances in Parasite Epidemiology and Population Genetics
A topical collection in Animals (ISSN 2076-2615). This collection belongs to the section "Veterinary Clinical Studies".
Viewed by 33093Editors
Interests: veterinary parasitology; zoonotic parasitic diseases; molecular epidemiology; genomics and genetics; parasite–host interactions
Topical Collection Information
Dear Colleagues,
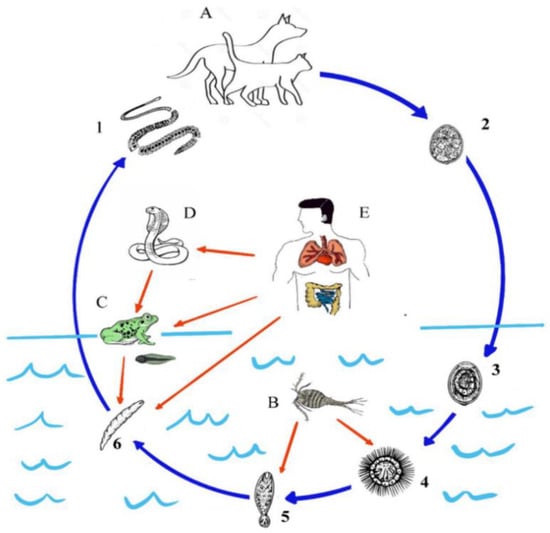
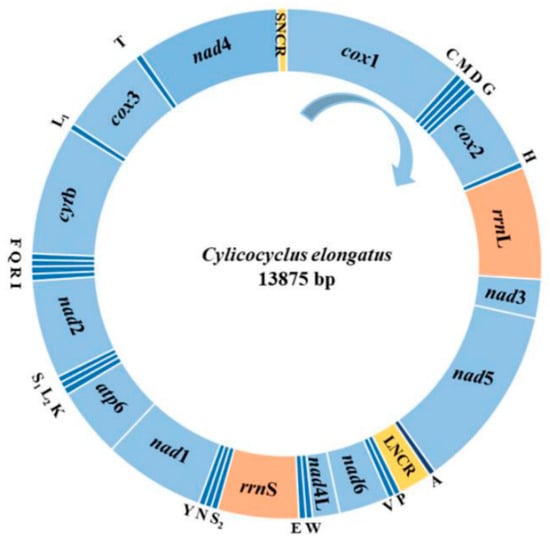
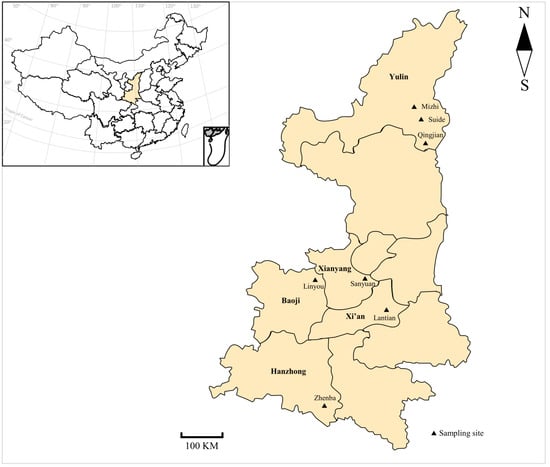
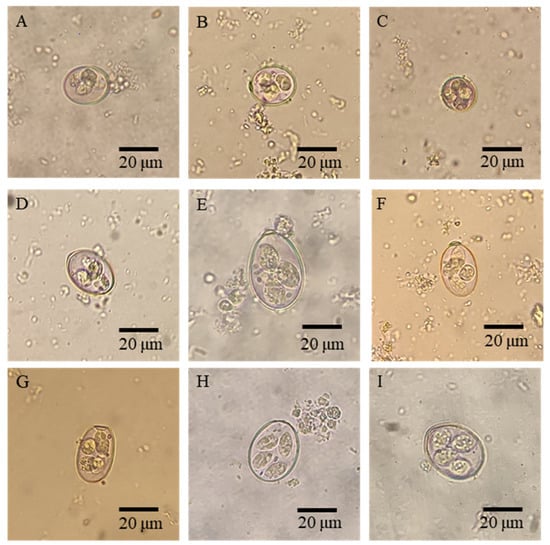
Intensive animal production, mass anthelmintic treatment, and climate change have significantly changed the transmission of parasitic diseases, the management of animals, and the wellbeing of humans. This Topical Collection focuses on the major advances in epidemiological and population genetic study of animal parasites. Epidemiological studies include etiology, surveillance and geographical spread of parasitic diseases using traditional examination and immunological, molecular or meta-analysis tools, as well as investigations of anthelmintic treatment effects and drug resistance in clinical trials for domestic and wild animals. Population genetic studies include genetic variants within parasite populations, genetic variants in response to environmental, host or anthelmintic stress revealed by genome sequencing and bioinformatic analysis using cutting-edge tools.
We welcome original research papers that provide a better understanding of the epidemiology, drug resistance and genetic variants of parasites driven by climate change, anthelmintic treatment and host immune responses. Areas of interest include epidemiological, population genetic, and drug resistance surveillances of parasites contributing to a better management and welfare of domestic and wild animals, as well as studies under one health concept preventing the transmission of parasites from animals to human beings.
Dr. Guangxu Ma
Dr. Yue Xie
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Animals is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2400 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- animal parasites
- helminths
- protozoan
- vector insects
- epidemiology
- population genetics