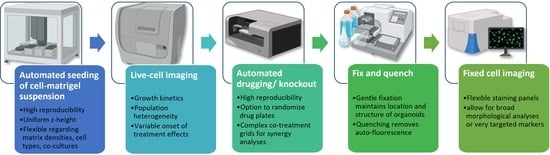
High-Throughput Live and Fixed Cell Imaging Method to Screen Matrigel-Embedded Organoids
Abstract
:1. Introduction
- (i)
- It changes the relative position of organoids in the well, which makes tracking of individual structures impossible.
- (ii)
- It can change the original morphology of organoids because of the loss of the supporting matrix, especially larger structures or cystic organoids.
- (iii)
- Organoids are likely to form clumps that are very hard to segment during image analysis, which makes single organoid analyses almost impossible.
2. Materials and Methods
2.1. Cell Culture and Spheroid Preparation
2.2. Live-Cell Image Acquisition
2.3. Fixing and Quenching
2.4. Multiplex Staining of Spheroids to Image Fixed Cells
2.5. Fixed-Cell Image Acquisition
2.6. Compound Screen
2.7. Image Analysis
2.8. Data Analysis
- (i)
- inactive features with low/no variance (=69 features removed).
- (ii)
- features containing NA, NaN, or Inf (=11 features removed).
3. Results
3.1. Optimisation of Fixation and Quenching
3.2. Optimized Method Allows Single Spheroid Tracking over Time and Post-Fixation
3.3. Optimisation of Screening Conditions to Allow for High Throughput
- (i)
- The 3D objects are distributed throughout the whole z-height of a large Matrigel dome and require lengthy autofocus in each well or even each individual field.
- (ii)
- The method is very specific to a certain cell type and cannot be generalized to a large range of cell types and Matrigel concentrations.
- (iii)
- After finding the first focal plane, the assay requires a large z-stack in each field.
- (iv)
- The distribution of the 3D objects in the well is not even or too sparse, which requires dozens of fields when imaging at higher magnifications—because many fields will be empty.
3.4. High-Content Screen with Optimized Fixing and Staining Method
3.4.1. Results before and after Fixation and Across Imaging Platforms
3.4.2. Unsupervised Feature Reduction
3.4.3. Active Compound Clustering
- Median_Nuclei_Intensity_IntegratedIntensity,
- Median_Nuclei_RadialDistribution_FracAtD_2of4,
- Median_Nuclei_Texture_InfoMeas1_10_00_256 (measure of the total amount of information contained within a region of pixels derived from the recurring spatial relationship between specific intensity values),
- StDev_Nuclei_Intensity_IntegratedIntensity

4. Discussion
- (a)
- flexible in the cell type that can be used and depth of information that can be extracted, ranging from basic live-cell readouts to single-cell intra-organoid heterogeneity;
- (b)
- economical in the use of consumables (Matrigel), patient/cell material, and imaging time;
- (c)
- instrument agnostic in the required screening equipment (liquid-handling, microscope platform) as well as a publicly available image analysis software.

4.1. Flexible
4.2. Economical
4.3. Instrument Agnostic
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carpenter, A.E. Image-based chemical screening. Nat. Chem. Biol. 2007, 3, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Lukonin, I.; Zinner, M.; Liberali, P. Organoids in image-based phenotypic chemical screens. Exp. Mol. Med. 2021, 53, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Birgersdotter, A.; Sandberg, R.; Ernberg, I. Gene expression perturbation in vitro—A growing case for three-dimensional (3D) culture systems. Semin. Cancer Biol. 2005, 15, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, W.R.; Foster, A.J.; Vogt, J.; Summers, C.; Middleton, B.; Pilling, M.A.; Shienson, D.; Kijanska, M.; Strobel, S.; Kelm, J.M.; et al. Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch. Toxicol. 2017, 91, 2849–2863. [Google Scholar] [CrossRef]
- Kenny, H.A.; Lal-Nag, M.; White, E.A.; Shen, M.; Chiang, C.Y.; Mitra, A.K.; Zhang, Y.; Curtis, M.; Schryver, E.M.; Bettis, S.; et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat. Commun. 2015, 6, 6220. [Google Scholar] [CrossRef] [Green Version]
- Beghin, A.; Grenci, G.; Sahni, G.; Guo, S.; Rajendiran, H.; Delaire, T.; Mohamad Raffi, S.B.; Blanc, D.; de Mets, R.; Ong, H.T.; et al. Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat. Methods 2022, 19, 881–892. [Google Scholar] [CrossRef]
- Choo, N.; Ramm, S.; Luu, J.; Winter, J.M.; Selth, L.A.; Dwyer, A.R.; Frydenberg, M.; Grummet, J.; Sandhu, S.; Hickey, T.E.; et al. High-Throughput Imaging Assay for Drug Screening of 3D Prostate Cancer Organoids. SLAS Discov. 2021, 26, 1107–1124. [Google Scholar] [CrossRef]
- Bray, M.A.; Singh, S.; Han, H.; Davis, C.T.; Borgeson, B.; Hartland, C.; Kost-Alimova, M.; Gustafsdottir, S.M.; Gibson, C.C.; Carpenter, A.E. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 2016, 11, 1757–1774. [Google Scholar] [CrossRef] [Green Version]
- Broguiere, N.; Isenmann, L.; Hirt, C.; Ringel, T.; Placzek, S.; Cavalli, E.; Ringnalda, F.; Villiger, L.; Züllig, R.; Lehmann, R.; et al. Growth of Epithelial Organoids in a Defined Hydrogel. Adv. Mater. 2018, 30, 1801621. [Google Scholar] [CrossRef]
- Boutin, M.E.; Voss, T.C.; Titus, S.A.; Cruz-Gutierrez, K.; Michael, S.; Ferrer, M. A high-throughput imaging and nuclear segmentation analysis protocol for cleared 3D culture models. Sci. Rep. 2018, 8, 11135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezakhani, S.; Gjorevski, N.; Lutolf, M.P. Extracellular matrix requirements for gastrointestinal organoid cultures. Biomaterials 2021, 276, 121020. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.H.; Kang, D.; Seo, S.J.; Jin, Y. Engineering the Extracellular Matrix for Organoid Culture. Int. J. Stem Cells 2022, 15, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Risbridger, G.P.; Clark, A.K.; Porter, L.H.; Toivanen, R.; Bakshi, A.; Lister, N.L.; Pook, D.; Pezaro, C.J.; Sandhu, S.; Keerthikumar, S.; et al. The MURAL collection of prostate cancer patient-derived xenografts enables discovery through preclinical models of uro-oncology. Nat. Commun. 2021, 12, 5049. [Google Scholar] [CrossRef]
- Hutz, J.E.; Nelson, T.; Wu, H.; McAllister, G.; Moutsatsos, I.; Jaeger, S.A.; Bandyopadhyay, S.; Nigsch, F.; Cornett, B.; Jenkins, J.L.; et al. The Multidimensional Perturbation Value: A Single Metric to Measure Similarity and Activity of Treatments in High-Throughput Multidimensional Screens. J. Biomol. Screen. 2012, 18, 367–377. [Google Scholar] [CrossRef] [Green Version]
- Clancy, B.; Cauller, L.J. Reduction of background autofluorescence in brain sections following immersion in sodium borohydride. J. Neurosci. Methods 1998, 83, 97–102. [Google Scholar] [CrossRef]
- Boland, M.V.; Markey, M.K.; Murphy, R.F. Automated recognition of patterns characteristic of subcellular structures in fluorescence microscopy images. Cytometry 1998, 33, 366–375. [Google Scholar] [CrossRef]
- Boutros, M.; Heigwer, F.; Laufer, C. Microscopy-Based High-Content Screening. Cell 2015, 163, 1314–1325. [Google Scholar] [CrossRef] [Green Version]
- Pegoraro, G.; Misteli, T. High-Throughput Imaging for the Discovery of Cellular Mechanisms of Disease. Trends Genet. 2017, 33, 604–615. [Google Scholar] [CrossRef]
- Badder, L.M.; Hollins, A.J.; Herpers, B.; Yan, K.; Ewan, K.B.; Thomas, M.; Shone, J.R.; Badder, D.A.; Naven, M.; Ashelford, K.E.; et al. 3D imaging of colorectal cancer organoids identifies responses to Tankyrase inhibitors. PLoS ONE 2020, 15, e0235319. [Google Scholar] [CrossRef]
- Bock, C.; Boutros, M.; Camp, J.G.; Clarke, L.; Clevers, H.; Knoblich, J.A.; Liberali, P.; Regev, A.; Rios, A.C.; Stegle, O.; et al. The Organoid Cell Atlas. Nat. Biotechnol. 2021, 39, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Ceder, S.; Eriksson, S.E.; Cheteh, E.H.; Dawar, S.; Corrales Benitez, M.; Bykov, V.J.N.; Fujihara, K.M.; Grandin, M.; Li, X.; Ramm, S.; et al. A thiol-bound drug reservoir enhances APR-246-induced mutant p53 tumor cell death. EMBO Mol. Med. 2021, 13, e10852. [Google Scholar] [CrossRef] [PubMed]
- Behrenbruch, C.; Foroutan, M.; Lind, P.; Smith, J.; Grandin, M.; Cooper, B.; Shembrey, C.; Ramm, S.; Cowley, K.; Nikolic, I.; et al. Targeting of TP53-independent cell cycle checkpoints overcomes FOLFOX resistance in Metastatic Colorectal Cancer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Betge, J.; Rindtorff, N.; Sauer, J.; Rauscher, B.; Dingert, C.; Gaitantzi, H.; Herweck, F.; Srour-Mhanna, K.; Miersch, T.; Valentini, E.; et al. The drug-induced phenotypic landscape of colorectal cancer organoids. Nat. Commun. 2022, 13, 3135. [Google Scholar] [CrossRef] [PubMed]
- Fujii, E.; Yamazaki, M.; Kawai, S.; Ohtani, Y.; Watanabe, T.; Kato, A.; Suzuki, M. A simple method for histopathological evaluation of organoids. J. Toxicol. Pathol. 2018, 31, 81–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenzie, A.T. Glutaraldehyde: A Review of Its Fixative Effects on Nucleic Acids, Proteins, Lipids, and Carbohydrates. [CrossRef]
- Kong, I.Y.; Trezise, S.; Light, A.; Todorovski, I.; Arnau, G.M.; Gadipally, S.; Yoannidis, D.; Simpson, K.J.; Dong, X.; Whitehead, L.; et al. Epigenetic modulators of B cell fate identified through coupled phenotype-transcriptome analysis. Cell Death Differ. 2022, 29, 2519–2530. [Google Scholar] [CrossRef]
- Lukonin, I.; Serra, D.; Challet Meylan, L.; Volkmann, K.; Baaten, J.; Zhao, R.; Meeusen, S.; Colman, K.; Maurer, F.; Stadler, M.B.; et al. Phenotypic landscape of intestinal organoid regeneration. Nature 2020, 586, 275–280. [Google Scholar] [CrossRef]
- Mergenthaler, P.; Hariharan, S.; Pemberton, J.M.; Lourenco, C.; Penn, L.Z.; Andrews, D.W. Rapid 3D phenotypic analysis of neurons and organoids using data-driven cell segmentation-free machine learning. PLoS Comput. Biol. 2021, 17, e1008630. [Google Scholar] [CrossRef]
- Hongisto, V.; Jernstrom, S.; Fey, V.; Mpindi, J.P.; Kleivi Sahlberg, K.; Kallioniemi, O.; Perala, M. High-throughput 3D screening reveals differences in drug sensitivities between culture models of JIMT1 breast cancer cells. PLoS ONE 2013, 8, e77232. [Google Scholar] [CrossRef]
- Du, Y.; Li, X.; Niu, Q.; Mo, X.; Qui, M.; Ma, T.; Kuo, C.J.; Fu, H. Development of a miniaturized 3D organoid culture platform for ultra-high-throughput screening. J. Mol. Cell Biol. 2020, 12, 630–643. [Google Scholar] [CrossRef]
- Engel, M.; Belfiore, L.; Aghaei, B.; Sutija, M. Enabling high throughput drug discovery in 3D cell cultures through a novel bioprinting workflow. SLAS Technol. 2022, 27, 32–38. [Google Scholar] [CrossRef] [PubMed]






| Pre-Treatment | Fixative | Concentration (v/v%) | Incubation Time (Minutes) |
|---|---|---|---|
| None | FA diluted in PBS | 2, 4 | 10 |
| Sucrose | FA diluted in PBS | 2, 4 | 1, 3, 5, 10 |
| None | Glutaraldehyde diluted in PBS | 0.1, 0.3, 0.5, 1 | 1, 3, 5, 10 |
| None | Glutaraldehyde diluted in PBS | 0.4 | 10 |
| Concentration (%) of Sodium Borohydride | Incubation Time | Incubation Temp. |
|---|---|---|
| 0.5 | 20 min | RT |
| 0.5 | 35 min | RT |
| 1, 3, 5 | 60 min | RT |
| 1, 1.5, 2 | 18 h | 4 °C |
| 0.7, 0.8, 0.9 | 18 h | 4 °C |
| 0.05, 0.2, 0.5 | 1, 4 h | 4 °C |
| Target | Reagents | Catalog Number | Stock | Final Dilution | Diluent | Incubation Time and Temp. |
|---|---|---|---|---|---|---|
| Mito-chondria | MitoTracker Deep Red dye | M22426 | 1 mM | 1:500 | Media | 2 h at 37 °C in incubator |
| Nuclei | DAPI | D9542 | 5 mg/mL | 1:1000 | 50 mM Tris pH 7.6 | 2 h at RT |
| Golgi | Alexa Fluor 594-WGA | W11262 | 1 mg/mL | 1: 00 | 50 mM Tris pH 7.6 | 2 h at RT |
| F-Actin cyto-skeleton | Rhodamine Phalloidin | 00027 | 200 Units/mL (~6.6 µM) | 1: 50 | 50 mM Tris pH 7.6 | 2 h at RT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramm, S.; Vary, R.; Gulati, T.; Luu, J.; Cowley, K.J.; Janes, M.S.; Radio, N.; Simpson, K.J. High-Throughput Live and Fixed Cell Imaging Method to Screen Matrigel-Embedded Organoids. Organoids 2023, 2, 1-19. https://doi.org/10.3390/organoids2010001
Ramm S, Vary R, Gulati T, Luu J, Cowley KJ, Janes MS, Radio N, Simpson KJ. High-Throughput Live and Fixed Cell Imaging Method to Screen Matrigel-Embedded Organoids. Organoids. 2023; 2(1):1-19. https://doi.org/10.3390/organoids2010001
Chicago/Turabian StyleRamm, Susanne, Robert Vary, Twishi Gulati, Jennii Luu, Karla J. Cowley, Michael S. Janes, Nicholas Radio, and Kaylene J. Simpson. 2023. "High-Throughput Live and Fixed Cell Imaging Method to Screen Matrigel-Embedded Organoids" Organoids 2, no. 1: 1-19. https://doi.org/10.3390/organoids2010001