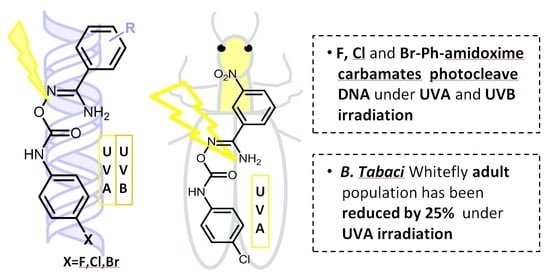
Exploration of the DNA Photocleavage Activity of O-Halo-phenyl Carbamoyl Amidoximes: Studies of the UVA-Induced Effects on a Major Crop Pest, the Whitefly Bemisia tabaci †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Chemical Compounds
- (Z)-N′-(4-chlorophenylcarbamoyloxy)-4-nitrobenzimidamide (16): reflux: 1 h; orange amorphous solid; mp: 196–198 °C (E.A./EtOH); yield: 91%; 1H−NMR (DMSO−d6, 500 MHz) δ 9.58 (s, 1H, NH), 8.31 (d, J = 8.5 Hz, 2H), 8.10 (d, J = 8.3 Hz, 2H), 7.57 (d, J = 8.5, 2H), 7.38 (d, J = 8.4 Hz, 2H), 7.09 (brs, 2H, NH2) ppm; 13C−NMR (DMSO−d6, 125 MHz) δ 154.2, 152.3, 148.7, 137.5, 137.4, 128.7, 128.2, 126.7, 123.5, 120.7 ppm; HRMS(ESI) m/z [M+Na]+: C14H11ClN4O4Na+, calc: 357.0361; found: 357.0361.
- (Z)-N′-(4-chlorophenylcarbamoyloxy)-3-nitrobenzimidamide (17): reflux: 1 h; yellow amorphous solid; mp: 210–212 °C (E.A.); yield: 89%; 1H−NMR (DMSO−d6, 500 MHz) δ 9.62 (s, 1H, NH), 8.63 (s, 1H), 8.36 (d, J = 7.8 Hz, 1H), 8.27 (d, J = 7.6 Hz, 1H), 7.78 (t, J = 7.9 Hz, 1H), 7.56 (d, J = 8.5 Hz, 2H), 7.37 (d, J = 8.5 Hz, 2H), 7.11 (brs, 2H, NH2) ppm; 13C−NMR (DMSO−d6, 125 MHz) δ 154.1, 152.4, 147.8, 137.6, 133.2, 132.9, 130.2, 128.7, 126.7, 125.3, 121.6, 120.7 ppm; HRMS(ESI) m/z [M+Na]+: C14H11ClN4O4Na+, calc: 357.0361; found: 357.0366.
- (Z)-N′-(4-chlorophenylcarbamoyloxy)-2-nitrobenzimidamide (18): reflux: 1 h; beige amorphous solid; mp: 142–145 °C (E.A./hex); yield: 50%; 1H−NMR (DMSO−d6, 500 MHz) δ 9.41 (s, 1H, NH), 8.08 (d, J = 8 Hz, 1H), 7.84 (t, J = 7.5 Hz, 1H), 7.78–7.70 (m, 2H), 7.53 (d, J = 8.7 Hz, 2H), 7.35 (d, J = 8.7 Hz, 2H), 7.08 (brs, 2H, NH2) ppm; 13C−NMR (DMSO−d6, 125 MHz) δ 154.8, 152.2, 148.4, 137.6, 133.4, 131.4, 131.3, 128.6, 126.7, 126.5, 124.2, 120.4 ppm; HRMS(ESI) m/z [M+Na]+: C14H11ClN4O4Na+, calc: 357.0361; found: 357.0367.
- (Z)-2-nitro-N′-(4-nitrophenylcarbamoyloxy)benzimidamide (19): reflux: 1 h; yellow amorphous solid; mp: 142–145 °C (EtOH/THF); yield: 75%; 1H−NMR (DMSO−d6, 500 MHz) δ 10.14 (s, 1H, NH), 9.32 (s, 1H, NH), 9.00 (s, 1H, NH), 8.15 (d, J = 8.5 Hz, 2H), 8.13 (d, J = 8.3 Hz, 1H), 7.87–7.78 (m, 3H), 7.73 (dt, J = 8.2, 1.3 Hz, 1H), 7.67 (d, J = 7.5 Hz, 1H) ppm; 13C−NMR (DMSO−d6, 125 MHz) δ 165.7, 162.3, 147.0, 146.9, 141.1, 133.9, 132.0, 130.7, 129.9, 125.0, 124.2, 117.7 ppm; HRMS(ESI) m/z [M+Na]+: C14H11N5O6Na+, calc: 368.0607; found: 368.0564.
- (Z)-N′-(4-methoxyphenylcarbamoyloxy)-2-nitrobenzimidamide (20): reflux: 1 h; pale yellow amorphous solid; mp: 129–131 °C (E.A./EtOH); yield: 82%; 1H−NMR (DMSO−d6, 500 MHz) δ 9.03 (s, 1H, NH), 8.08 (d, J = 8 Hz, 1H), 7.83 (t, J = 7.3 Hz, 1H), 7.79–7.70 (m, 2H), 7.39 (d, J = 8.9 Hz, 2H), 7.08 (brs, 2H, NH2), 6.88 (d, J = 9.0 Hz, 2H), 3.71 (s, 3H) ppm; 13C−NMR (DMSO−d6, 125 MHz) δ 155.2, 154.3, 152.6, 148.5, 133.4, 131.5, 131.4, 131.3, 126.8, 124.2, 120.7, 114.0, 55.2 ppm; HRMS(ESI) m/z [M+Na]+: C15H14N4O5Na+, calc: 353.0856; found: 353.0871.
- (Z)-N′-(4-chlorophenylcarbamoyloxy)benzimidamide (21): reflux: 1 h; White amorphous solid; mp: 155–156 °C (hex/E.A.); yield: 90%; 1H−NMR (CDCl3, 500 MHz) δ 8.67 (s, 1H, NH), 7.68 (d, J = 7.5 Hz, 2H), 7.54 (t, J = 7.3 Hz, 1H), 7.50–7.43 (m, 4H), 7.29 (d, J = 8.6 Hz, 2H), 5.34 (brs, 2H, NH2) ppm; 13C−NMR (CDCl3, 125 MHz) δ 154.8, 152.7, 136.0, 131.5, 130.9, 129.2, 129.2, 129.1, 126.7, 121.0 ppm; HRMS(ESI) m/z [M+Na]+: C14H12ClN3O2Na+, calc: 312.0510; found: 312.0510.
- (Z)-N′-(4-chlorophenylcarbamoyloxy)-4-methoxybenzimidamide (22): reflux: 1 h; White amorphous solid; mp: 175–177 °C (hex/E.A.); yield: 88%; 1H−NMR (CDCl3, 500 MHz) δ 8.68 (brs, 1H, NH), 7.62 (d, J = 8.7 Hz, 2H), 7.46 (d, J = 8.7 Hz, 2H), 7.29 (d, J = 8.7 Hz, 2H), 6.97 (d, J = 8.7 Hz, 2H), 5.27 (brs, 2H, NH2), 3.86 (s, 3H) ppm; 13C−NMR (CDCl3, 125 MHz) δ 162.2, 154.5, 152.8, 136.1, 129.2, 128.2, 123.0, 121.0, 114.4, 55.6 ppm; HRMS(ESI) m/z [M+Na]+: C15H14ClN3O3Na+, calc: 342.0616; found: 342.0613.
- (Z)-N′-(4-fluorophenylcarbamoyloxy)-4-methoxybenzimidamide (23): reflux: 3 h; White amorphous solid; mp: 179–181 °C (hex/E.A.); yield: 89%; 1H−NMR (DMSO−d6, 500 MHz) δ 9.04 (s, 1H, NH), 7.77 (d, J = 8.9 Hz, 2H), 7.55 (dd, J = 9.0, 5.0 Hz, 2H), 7.16 (t, J = 8.9 Hz, 2H), 6.99 (d, J = 8.9 Hz, 2H), 6.74 (brs, 2H, NH2), 3.81 (s, 3H) ppm; 13C−NMR (DMSO−d6, 125 MHz) δ 161.0, 158.0 (d, 1JC-F = 238 Hz), 155.2, 152.9, 134.9 (d, 4JC-F = 2.5 Hz), 128.3, 123.4, 122.1 (d, 3JC-F = 7.8 Hz), 115.3 (d, 2JC-F = 28 Hz), 113.7, 55.4 ppm; HRMS(ESI) m/z [M+Na]+: C15H14FN3O3Na+, calc: 326.0917; found: 326.0910.
- (Z)-N′-(4-bromophenylcarbamoyloxy)-4-methoxybenzimidamide (24): reflux: 1 h; White amorphous solid; mp: 150–152 °C (hex/E.A.); yield: 80%; 1H−NMR (CDCl3, DMSO−d6, calibration on CDCl3, 500 MHz) δ 8.70 (s, 1H, NH), 7.58 (d, J = 8.7 Hz, 2H), 7.35 (s, 4H), 6.88 (d, J = 8.7 Hz, 2H), 5.63 (brs, 2H, NH2), 3.78 (s, 3H) ppm; 13C−NMR (CDCl3, DMSO−d6, calibration on CDCl3, 125 MHz) δ 161.8, 154.8, 152.7, 136.7, 131.8, 128.1, 123.0, 121.1, 116.34, 113.9, 55.4 ppm; HRMS(ESI) m/z [M+Na]+: C15H14BrN3O3Na+, calc: 386.0116; found: 386.0111.
- (Z)-N′-(4-iodophenylcarbamoyloxy)-4-methoxybenzimidamide (25): reflux: 2 h; pale white amorphous solid; mp: 180–182 °C (hex/E.A.); yield: 85%; 1H−NMR (CDCl3, 500 MHz) δ 8.67 (s, 1H, NH), 7.62 (d, J = 8.6 Hz, 2H), 7.61 (d, J = 8.6 Hz, 2H), 7.30 (d, J = 8.6 Hz, 2H), 6.97 (d, J = 8.7 Hz, 2H), 5.26 (brs, 2H, NH2), 3.86 (s, 3H) ppm; 13C−NMR (CDCl3, 125 MHz) δ 162.2, 154.5, 152.6, 138.1, 137.3, 128.2, 123.0, 121.6, 114.5, 87.2, 55.6 ppm; HRMS(ESI) m/z [M+Na]+: C15H14IN3O3Na+, calc: 433.9972; found: 433.9970.
- (Z)-N′-(3-chlorophenylcarbamoyloxy)-4-methoxybenzimidamide (26): reflux: 3 h; White amorphous solid; mp: 162–165 °C (E.A./hex); yield: 90%; 1H−NMR (CDCl3, DMSO−d6, calibration on CDCl3, 500 MHz) δ 8.73 (s, 1H, NH), 7.41 (d, J = 8.5 Hz, 2H), 7.37 (s, 1H), 7.07 (d, J = 8.2 Hz, 1H), 6.94 (t, J = 8.0 Hz, 1H), 6.73 (d, J = 8.0 Hz, 1H), 6.64 (d, J = 8.5 Hz, 2H), 5.85 (brs, 2H, NH2), 3.54 (s, 3H) ppm; 13C−NMR (CDCl3, DMSO−d6, calibration on CDCl3, 125 MHz) δ 161.0, 154.6, 152.1, 138.7, 133.7, 129.4, 127.8, 122.8, 122.6, 118.6, 116.9, 113.3, 54.8 ppm; HRMS(ESI) m/z [M+Na]+: C15H14ClN3O3Na+, calc: 342.0616; found: 342.0621.
- (Z)-N′-(2-chlorophenylcarbamoyloxy)-4-methoxybenzimidamide (27): reflux: 6 h; White amorphous solid; mp: 133–135 °C (E.A./hex); yield: 82%; 1H−NMR (CDCl3, 500 MHz) δ 9.47 (s, 1H, NH), 8.30 (d, J = 8.2 Hz, 1H), 7.66 (d, J = 8.4 Hz, 2H), 7.38 (d, J = 8.0 Hz, 1H), 7.30 (t, J = 7.6 Hz, 1H), 7.03 (t, J = 7.6 Hz, 1H), 6.97 (d, J = 8.5 Hz, 2H), 5.29 (brs, 2H, NH2), 3.85 (s, 3H) ppm; 13C−NMR (CDCl3, 125 MHz) δ 162.1, 154.1, 152.6, 134.6, 129.2, 127.9, 127.9, 124.2, 123.2, 122.8, 120.5, 114.4, 55.5 ppm; HRMS(ESI) m/z [M+Na]+: C15H14ClN3O3Na+, calc: 342.0616; found: 342.0619.
- (Z)-N′-(((3,4-dichlorophenyl)carbamoyl)oxy)-4-methoxybenzimidamide (28): reflux: 3 h; White amorphous solid; mp: 153–155 °C (E.A./hex); yield: 80%; 1H−NMR (CDCl3, 500 MHz) δ 8.72 (s, 1H, NH), 7.72 (s, 1H), 7.61 (d, J = 8.7 Hz, 2H), 7.37 (brs, 2H), 6.97 (d, J = 8.7 Hz, 2H), 5.23 (brs, 2H, NH2), 3.86 (s, 3H) ppm; 13C−NMR (CDCl3, 125 MHz) δ 162.2, 154.7, 152.5, 137.1, 133.0, 130.7, 128.2, 127.3, 122.8, 121.3, 118.9, 114.5, 55.6 ppm; HRMS(ESI) m/z [M+Na]+: C15H13Cl2N3O3Na+, calc: 376.0226; found: 376.0223.
- (Z)-N′-(2,4-dichlorophenylcarbamoyloxy)-4-methoxybenzimidamide (29): reflux: 3 h; White amorphous solid; mp: 154–157 °C (E.A./hex); yield: 79%; 1H−NMR (CDCl3, 500 MHz) δ 9.40 (s, 1H, NH), 8.15 (d, J = 9.0 Hz, 1H), 7.70 (d, J = 8.9 Hz, 2H), 7.48 (d, J = 2.2 Hz, 1H), 7.31 (dd, J = 8.9, 2.2 Hz, 1H), 6.94 (d, J = 8.8 Hz, 2H), 6.85 (brs, 2H, NH2), 3.79 (s, 3H) ppm; 13C−NMR (CDCl3, 125 MHz) δ 161.0, 154.7, 151.8, 133.5, 128.3, 127.8, 127.5, 127.4, 123.3, 122.7, 121.2, 113.5, 55.0 ppm; HRMS(ESI) m/z [M+Na]+: C15H13Cl2N3O3Na+, calc: 376.0226; found: 376.0224.
2.2. Supercoiled Circular pB322 DNA Photo-Cleavage Experiments
2.3. Toxicological Bioassays of Bemisia tabaci
2.3.1. Bioassays of Bemisia tabaci
2.3.2. Data Analysis and Statistics
3. Results and Discussion
3.1. Chemistry
3.2. DNA Photocleavage Experiments
3.3. Pesticidal Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McBurney, R.T.; Walton, J.C. Dissociation or Cyclization: Options for a Triad of Radicals Released from Oxime Carbamates. J. Am. Chem. Soc. 2013, 135, 7349–7354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsunooka, M.; Suyama, K.; Okamura, H.; Shirai, M. Development of Photoacid and Photobase Generators as the Key Materials for Design of Novel Photopolymers. J. Photopolym. Sci. Technol. 2006, 19, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Yang, Y.; Yang, H.; Han, F.; Li, Z. Thioxanthone-Based Amidine: An Efficient Nonionic Photobase Generator for Thiol-Based Click Polymerization under Visible LED Light. Prog. Org. Coat. 2020, 148, 105842. [Google Scholar] [CrossRef]
- Huang, L.; Xie, G.; Yang, J. A Fluorinated Photobase Generator with UVA Sensitive for Surface Oxygen Inhibition. Prog. Org. Coat. 2020, 143, 105604. [Google Scholar] [CrossRef]
- Zhang, X.; Xi, W.; Gao, G.; Wang, X.; Stansbury, J.W.; Bowman, C.N. O-Nitrobenzyl-Based Photobase Generators: Efficient Photoinitiators for Visible-Light Induced Thiol-Michael Addition Photopolymerization. ACS Macro Lett. 2018, 7, 852–857. [Google Scholar] [CrossRef]
- Terada, K.; Furutani, M.; Arimitsu, K. Development of Photobase Generators Liberating Radicals as Well as Bases and Their Application to Hardcoating Materials. J. Photopolym. Sci. Technol. 2018, 31, 493–496. [Google Scholar] [CrossRef] [Green Version]
- Arimitsu, K.; Oguri, A.; Furutani, M. 365 Nm-Light-Sensitive Photobase Generators Derived from Trans-o-Coumaric Acid. Mater. Lett. 2015, 140, 92–94. [Google Scholar] [CrossRef]
- Zivic, N.; Kuroishi, P.K.; Dumur, F.; Gigmes, D.; Dove, A.P.; Sardon, H. Recent Advances and Challenges in the Design of Organic Photoacid and Photobase Generators for Polymerizations. Angew. Chem. Int. Ed. 2019, 58, 10410–10422. [Google Scholar] [CrossRef]
- De Pariza, X.L.; Jara, E.C.; Zivic, N.; Ruipérez, F.; Long, T.E.; Sardon, H. Novel Imino- and Aryl-Sulfonate Based Photoacid Generators for the Cationic Ring-Opening Polymerization of ε-Caprolactone. Polym. Chem. 2021, 12, 4035–4042. [Google Scholar] [CrossRef]
- Lalevée, J.; Allonas, X.; Fouassier, J.P.; Tachi, H.; Izumitani, A.; Shirai, M.; Tsunooka, M. Investigation of the Photochemical Properties of an Important Class of Photobase Generators: The O-Acyloximes. J. Photochem. Photobiol. A Chem. 2002, 151, 27–37. [Google Scholar] [CrossRef]
- Walton, J.C. Functionalised Oximes: Emergent Precursors for Carbon-, Nitrogen- and Oxygen-Centred Radicals. Molecules 2016, 21, 63. [Google Scholar] [CrossRef] [Green Version]
- Alonso, R.; Caballero, A.; Campos, P.J.; Rodríguez, M.A. Photochemistry of Acyloximes: Synthesis of Heterocycles and Natural Products. Tetrahedron 2010, 66, 8828–8831. [Google Scholar] [CrossRef]
- Alonso, R.; Campos, P.J.; Rodríguez, M.A.; Sampedro, D. Photocyclization of Iminyl Radicals: Theoretical Study and Photochemical Aspects. J. Org. Chem. 2008, 73, 2234–2239. [Google Scholar] [CrossRef] [PubMed]
- Van Der Westhuyzen, R.; Mabhula, A.; Njaria, P.M.; Müller, R.; Muhunga, D.N.; Taylor, D.; Lawrence, N.; Njoroge, M.; Brunschwig, C.; Moosa, A.; et al. Benzoheterocyclic Oxime Carbamates Active against Mycobacterium Tuberculosis: Synthesis, Structure-Activity Relationship, Metabolism, and Biology Triaging. J. Med. Chem. 2021, 64, 9444–9457. [Google Scholar] [CrossRef] [PubMed]
- Keurulainen, L.; Heiskari, M.; Nenonen, S.; Nasereddin, A.; Kopelyanskiy, D.; Leino, T.O.; Yli-Kauhaluoma, J.; Jaffe, C.L.; Kiuru, P. Synthesis of Carboxyimidamide-Substituted Benzo[c][1,2,5]Oxadiazoles and Their Analogs, and Evaluation of Biological Activity against Leishmania Donovani. MedChemComm 2015, 6, 1673–1678. [Google Scholar] [CrossRef] [Green Version]
- Ke, S.; Yang, Z.; Shi, L.; Liu, M.; Liang, Y.; Wang, K.; Long, T.; Jiang, A.; Zhang, Z. Oxime Carbamates as Potential Insecticidal Agents: Design, Synthesis and Biological Evaluation. Lett. Drug Des. Discov. 2015, 12, 771–777. [Google Scholar] [CrossRef]
- Sun, M.; Xu, W.; Zhang, W.; Guang, C.; Mu, W. Microbial Elimination of Carbamate Pesticides: Specific Strains and Promising Enzymes. Appl. Microbiol. Biotechnol. 2022, 106, 5973–5986. [Google Scholar] [CrossRef]
- Malhotra, H.; Kaur, S.; Phale, P.S. Conserved Metabolic and Evolutionary Themes in Microbial Degradation of Carbamate Pesticides. Front. Microbiol. 2021, 12, 648868. [Google Scholar] [CrossRef]
- Bhatt, P.; Zhou, X.; Huang, Y.; Zhang, W.; Chen, S. Characterization of the Role of Esterases in the Biodegradation of Organophosphate, Carbamate, and Pyrethroid Pesticides. J. Hazard. Mater. 2021, 411, 125026. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Nong, S.; Bai, Z.; Han, N.; Wu, Q.; Huang, Z.; Ding, J. Display of a Novel Carboxylesterase CarCby on Escherichia Coli Cell Surface for Carbaryl Pesticide Bioremediation. Microb. Cell Factories 2022, 21, 97. [Google Scholar] [CrossRef]
- Moreira, S.; Silva, R.; Carrageta, D.F.; Alves, M.G.; Seco-Rovira, V.; Oliveira, P.F.; de Lourdes Pereira, M. Carbamate Pesticides: Shedding Light on Their Impact on the Male Reproductive System. Int. J. Mol. Sci. 2022, 23, 8206. [Google Scholar] [CrossRef]
- Lima, A.R.; Dias, L.D.; Garbuio, M.; Inada, N.M.; Bagnato, V.S. A Look at Photodynamic Inactivation as a Tool for Pests and Vector-Borne Diseases Control. Laser Phys. Lett. 2022, 19, 025601. [Google Scholar] [CrossRef]
- Ambrosini, V.; Issawi, M.; Sol, V.; Riou, C. Photodynamic Inactivation of Botrytis Cinerea by an Anionic Porphyrin: An Alternative Pest Management of Grapevine. Sci. Rep. 2020, 10, 17438. [Google Scholar] [CrossRef] [PubMed]
- Losi, A.; Gärtner, W. A Light Life Together: Photosensing in the Plant Microbiota. Photochem. Photobiol. Sci. 2021, 20, 451–473. [Google Scholar] [CrossRef]
- Stracke, F.; Heupel, M.; Thiel, E. Singlet Molecular Oxygen Photosensitized by Rhodamine Dyes: Correlation with Photophysical Properties of the Sensitizers. J. Photochem. Photobiol. A Chem. 1999, 126, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Jeon, R.; Wender, P.A. Photocleavage of DNA by 4′-Bromoacetophenone Analogs. Arch. Pharmacal Res. 2001, 24, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yuan, G.; Hu, J. Synthesis and Photo-Induced DNA Cleaving Activities of Conjugates of Halophenyl Derivative and Polyamide Containing N-Methylimidazoles. J. Photochem. Photobiol. B Biol. 2006, 82, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, A.; Balalas, T.; Mitrakas, A.; Vrazas, V.; Katsani, K.R.; Koumbis, A.E.; Koukourakis, M.I.; Litinas, K.E.; Fylaktakidou, K.C. 6-Nitro-Quinazolin–4(3H)–one Exhibits Photodynamic Effects and Photodegrades Human Melanoma Cell Lines. A Study on the Photoreactivity of Simple Quinazolin–4(3H)–ones. Photochem. Photobiol. 2021, 97, 826–836. [Google Scholar] [CrossRef]
- Qian, X.; Yao, W.; Chen, G.; Huang, X.; Mao, P. N-Aroyloxynaphthalimides as Novel Highly Efficient DNA Photocleavers: Substituent Effects. Tetrahedron Lett. 2001, 42, 6175–6178. [Google Scholar] [CrossRef]
- Chowdhury, N.; Dutta, S.; Dasgupta, S.; Singh, N.D.P.; Baidya, M.; Ghosh, S.K. Synthesis, Photophysical, Photochemical, DNA Cleavage/Binding and Cytotoxic Properties of Pyrene Oxime Ester Conjugates. Photochem. Photobiol. Sci. 2012, 11, 1239–1250. [Google Scholar] [CrossRef]
- Hwu, J.R.; Tsay, S.C.; Hong, S.C.; Hsu, M.H.; Liu, C.F.; Chou, S.S.P. Relationship between Structure of Conjugated Oxime Esters and Their Ability to Cleave DNA. Bioconjug. Chem. 2013, 24, 1778–1783. [Google Scholar] [CrossRef]
- Bindu, P.J.; Mahadevan, K.M.; Satyanarayan, N.D.; Naik, T.R.R. Synthesis and DNA Cleavage Studies of Novel Quinoline Oxime Esters. Bioorganic Med. Chem. Lett. 2012, 22, 898–900. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, X.; Qian, X.; Yao, W. N-Aroyloxylthioxo-Naphthalimides as DNA Photocleavers of Aroyloxyl Oxygen Radicals: Synthesis, Evaluation, and Substituents’ Effect. Bioorg. Med. Chem. 2004, 12, 2335–2341. [Google Scholar] [CrossRef]
- Karamtzioti, P.; Papastergiou, A.; Stefanakis, J.G.; Koumbis, A.E.; Anastasiou, I.; Koffa, M.; Fylaktakidou, K.C. O-Benzoyl Pyridine Aldoxime and Amidoxime Derivatives: Novel Efficient DNA Photo-Cleavage Agents. MedChemComm 2015, 6, 719–726. [Google Scholar] [CrossRef]
- Pasolli, M.; Dafnopoulos, K.; Andreou, N.P.; Gritzapis, P.S.; Koffa, M.; Koumbis, A.E.; Psomas, G.; Fylaktakidou, K.C. Pyridine and P-Nitrophenyl Oxime Esters with Possible Photochemotherapeutic Activity: Synthesis, DNA Photocleavage and DNA Binding Studies. Molecules 2016, 21, 864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreou, N.P.; Dafnopoulos, K.; Tortopidis, C.; Koumbis, A.E.; Koffa, M.; Psomas, G.; Fylaktakidou, K.C. Alkyl and Aryl Sulfonyl P-Pyridine Ethanone Oximes Are Efficient DNA Photo-Cleavage Agents. J. Photochem. Photobiol. B Biol. 2016, 158, 30–38. [Google Scholar] [CrossRef]
- Papastergiou, A.; Perontsis, S.; Gritzapis, P.; Koumbis, A.E.; Koffa, M.; Psomas, G.; Fylaktakidou, K.C. Evaluation of O-Alkyl and Aryl Sulfonyl Aromatic and Heteroaromatic Amidoximes as Novel Potent DNA Photo-Cleavers. Photochem. Photobiol. Sci. 2016, 15, 351–360. [Google Scholar] [CrossRef]
- Gritzapis, P.S.; Varras, P.C.; Andreou, N.P.; Katsani, K.R.; Dafnopoulos, K.; Psomas, G.; Peitsinis, Z.V.; Koumbis, A.E.; Fylaktakidou, K.C. P-Pyridinyl Oxime Carbamates: Synthesis, DNA Binding, DNA Photocleaving Activity and Theoretical Photodegradation Studies. Beilstein J. Org. Chem. 2020, 16, 337–350. [Google Scholar] [CrossRef] [Green Version]
- Siragusa, M.; Lentini, M.; Schepis, C. Agminated Lentiginosis in a Patient with Ring Chromosome 21. Eur. J. Dermatol. 2012, 22, 801–803. [Google Scholar] [CrossRef]
- Lehmann, P. UV-B-Therapie von Hautkrankheiten: Ein Update Zu Indikationen, Wirksamkeit, Nebenwirkungen Und Durchführung. Aktuelle Derm. 2018, 44, 15–18, In German. [Google Scholar] [CrossRef]
- Bala, H.R.; Khan, S.; Chong, A.H. Two Cases of Generalised Granuloma Annulare Successfully Treated with Acitretin and NB UVB Therapy. Australas. J. Dermatol. 2016, 57, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yi, X.; Li, Y.; Ding, Y. Efficacy Assessment of UVA1 and Narrowband UVB for Treatment of Scalp Psoriasis. Lasers Med. Sci. 2018, 33, 1979–1982. [Google Scholar] [CrossRef] [PubMed]
- Coricovac, D.; Farcas, C.; Nica, C.; Pinzaru, I.; Simu, S.; Stoian, D.; Soica, C.; Proks, M.; Avram, S.; Navolan, D.; et al. Ethinylestradiol and Levonorgestrel as Active Agents in Normal Skin, and Pathological Conditions Induced by UVB Exposure: In Vitro and in Ovo Assessments. Int. J. Mol. Sci. 2018, 19, 3600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zabolinejad, N.; Maleki, M.; Salehi, M.; Ashrafi, Z.; Molkara, S.; Layegh, P. Psoralen and Narrowband UVB Combination Provides Higher Efficacy in Treating Vitiligo Compared with Narrowband UVB Alone: A Randomised Clinical Trial. Australas. J. Dermatol. 2020, 61, e65–e69. [Google Scholar] [CrossRef] [PubMed]
- Al Salman, M.; Ghiasi, M.; Farid, A.S.; Taraz, M.; Azizpour, A.; Mahmoudi, H. Oral Simvastatin Combined with Narrowband UVB for the Treatment of Psoriasis: A Randomized Controlled Trial. Dermatol. Ther. 2021, 34, e15075. [Google Scholar] [CrossRef]
- Ara, S.; Mowla, M.R.; Alam, M.; Khan, I. Efficacy of Oral Methotrexate (MTX) Monotherapy vs. Oral MTX plus Narrowband Ultraviolet Light B Phototherapy in Palmoplantar Psoriasis. Dermatol. Ther. 2020, 33, e13486. [Google Scholar] [CrossRef]
- Martin, J.H.; Mifsud, D.; Rapisarda, C. The Whiteflies (Hemiptera: Aleyrodidae) of Europe and the Mediterranean Basin. Bull. Entomol. Res. 2000, 90, 407–448. [Google Scholar] [CrossRef] [Green Version]
- Bedford, I.D.; Briddon, R.W.; Brown, J.K.; Rosell, R.C.; Markham, P.G. Geminivirus Transmission and Biological Characterisation of Bemisia Tabaci (Gennadius) Biotypes from Different Geographic Regions. Ann. Appl. Biol. 1994, 125, 311–325. [Google Scholar] [CrossRef]
- Oliveira, M.R.V.; Anderson, P.; Oliveira, M.R.V.; Henneberry, T.J.; Anderson, P. History, Current Status, and Collaborative Research Projects for Bemisia Tabaci. Crop Prot. 2001, 20, 709–723. [Google Scholar] [CrossRef] [Green Version]
- Roditakis, E.; Roditakis, N.E.; Tsagkarakou, A. Insecticide Resistance in Bemisia Tabaci (Homoptera: Aleyrodidae) Populations from Crete. Pest Manag. Sci. 2005, 61, 577–582. [Google Scholar] [CrossRef]
- Sparks, T.C.; Nauen, R. IRAC: Mode of Action Classification and Insecticide Resistance Management. Pestic. Biochem. Physiol. 2015, 121, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Roditakis, E.; Grispou, M.; Morou, E.; Kristoffersen, J.B.; Roditakis, N.; Nauen, R.; Vontas, J.; Tsagkarakou, A. Current Status of Insecticide Resistance in Q Biotype Bemisia Tabaci Populations from Crete. Pest Manag. Sci. 2009, 65, 313–322. [Google Scholar] [CrossRef]
- Bielza, P.; Moreno, I.; Belando, A.; Grávalos, C.; Izquierdo, J.; Nauen, R. Spiromesifen and Spirotetramat Resistance in Field Populations of Bemisia Tabaci Gennadius in Spain. Pest Manag. Sci. 2019, 75, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Horowitz, A.R.; Ghanim, M.; Roditakis, E.; Nauen, R.; Ishaaya, I. Insecticide Resistance and Its Management in Bemisia Tabaci Species. J. Pest Sci. 2020, 93, 893–910. [Google Scholar] [CrossRef]
- Cai, J.; Wei, H.; Hong, K.H.; Wu, X.; Zong, X.; Cao, M.; Wang, P.; Li, L.; Sun, C.; Chen, B.; et al. Discovery, Bioactivity and Docking Simulation of Vorinostat Analogues Containing 1,2,4-Oxadiazole Moiety as Potent Histone Deacetylase Inhibitors and Antitumor Agents. Bioorganic Med. Chem. 2015, 23, 3457–3471. [Google Scholar] [CrossRef] [PubMed]
- Vörös, A.; Mucsi, Z.; Baán, Z.; Timári, G.; Hermecz, I.; Mizsey, P.; Finta, Z. An Experimental and Theoretical Study of Reaction Mechanisms between Nitriles and Hydroxylamine. Org. Biomol. Chem. 2014, 12, 8036–8047. [Google Scholar] [CrossRef] [PubMed]
- Mettu, A.; Talla, V.; Bajaj, D.M.; Subhashini, N.J.P. Design, Synthesis, and Molecular Docking Studies of Novel Pyrazolyl 2-Aminopyrimidine Derivatives as HSP90 Inhibitors. Arch. Pharm. Weinh. 2019, 352, 1900063. [Google Scholar] [CrossRef] [PubMed]
- IRAC Susceptibility Test Methods Series – Method No: 015. 2009. Available online: https://irac-online.org/methods/trialeurodes-vaporariorum-bemisia-tabaci-adult/ (accessed on 31 March 2023).
- IRAC Susceptibility Test Methods Series – Method No: 016. 2009. Available online: https://irac-online.org/methods/trialeurodes-vaporariorum-bemisia-tabaci-nymphs/ (accessed on 31 March 2023).
- Abbott, W.S. The Value of the Dry Substitutes for Liquid Lime. J. Econ. Entomol. 1925, 18, 265–267. [Google Scholar] [CrossRef]
- Yang, N.; Xie, W.; Jones, C.M.; Bass, C.; Jiao, X.; Yang, X.; Liu, B.; Li, R.; Zhang, Y. Transcriptome Profiling of the Whitefly Bemisia Tabaci Reveals Stage-Specific Gene Expression Signatures for Thiamethoxam Resistance. Insect Mol. Biol. 2013, 22, 485–496. [Google Scholar] [CrossRef] [Green Version]
- Nauen, R.; Bielza, P.; Denholm, I.; Gorman, K. Age-Specific Expression of Resistance to a Neonicotinoid Insecticide in the Whitefly Bemisia Tabaci. Pest Manag. Sci. 2008, 63, 1100–1106. [Google Scholar] [CrossRef]
- Jones, C.M.; Daniels, M.; Andrews, M.; Slater, R.; Lind, R.J.; Gorman, K.; Williamson, M.S.; Denholm, I. Age-Specific Expression of a P450 Monooxygenase (CYP6CM1) Correlates with Neonicotinoid Resistance in Bemisia Tabaci. Pestic. Biochem. Physiol. 2011, 101, 53–58. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panagopoulos, A.; Alipranti, K.; Mylona, K.; Paisidis, P.; Rizos, S.; Koumbis, A.E.; Roditakis, E.; Fylaktakidou, K.C. Exploration of the DNA Photocleavage Activity of O-Halo-phenyl Carbamoyl Amidoximes: Studies of the UVA-Induced Effects on a Major Crop Pest, the Whitefly Bemisia tabaci. DNA 2023, 3, 85-100. https://doi.org/10.3390/dna3020006
Panagopoulos A, Alipranti K, Mylona K, Paisidis P, Rizos S, Koumbis AE, Roditakis E, Fylaktakidou KC. Exploration of the DNA Photocleavage Activity of O-Halo-phenyl Carbamoyl Amidoximes: Studies of the UVA-Induced Effects on a Major Crop Pest, the Whitefly Bemisia tabaci. DNA. 2023; 3(2):85-100. https://doi.org/10.3390/dna3020006
Chicago/Turabian StylePanagopoulos, Anastasios, Konstantina Alipranti, Kyriaki Mylona, Polinikis Paisidis, Stergios Rizos, Alexandros E. Koumbis, Emmanouil Roditakis, and Konstantina C. Fylaktakidou. 2023. "Exploration of the DNA Photocleavage Activity of O-Halo-phenyl Carbamoyl Amidoximes: Studies of the UVA-Induced Effects on a Major Crop Pest, the Whitefly Bemisia tabaci" DNA 3, no. 2: 85-100. https://doi.org/10.3390/dna3020006