Size- and Stereochemistry-Dependent Transcriptional Bypass of DNA Alkyl Phosphotriester Adducts in Mammalian Cells
Abstract
:1. Introduction
2. Experimental Procedures
2.1. Construction of Transcription Templates
2.2. Cellular Transcription, RNA Extraction and Amplification
2.3. Restriction Digestion and Polyacrylamide Gel Electrophoresis (PAGE) Analysis
2.4. LC-MS/MS Analysis
3. Results
3.1. Effects of Alkyl-PTE Lesions on Transcription in Mammalian Cells
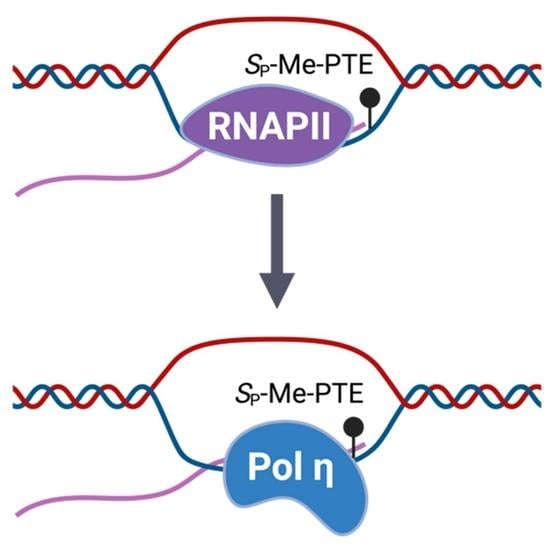
3.2. Pol η Promotes the Transcription of SP-Me-Alkyl-PTE Lesions in Mammalian Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Friedberg, E.C.; Walker, G.C.; Siede, W.; Wood, R.D.; Schultz, R.A.; Ellenberger, T. DNA Repair and Mutagenesis; ASM Press: Washington, DC, USA, 2006. [Google Scholar]
- Singer, B.; Grunberger, D. Molecular Biology of Mutagens and Carcinogens; Plenum Press: New York, NY, USA; London, UK, 1983. [Google Scholar]
- Fu, D.; Calvo, J.A.; Samson, L.D. Balancing Repair and Tolerance of DNA Damage Caused by Alkylating Agents. Nat. Rev. Cancer 2012, 12, 104–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedgwick, B.; Bates, P.A.; Paik, J.; Jacobs, S.C.; Lindahl, T. Repair of Alkylated DNA: Recent Advances. DNA Repair Amst. 2007, 6, 429–442. [Google Scholar] [CrossRef]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA Repair Pathways as Targets for Cancer Therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef]
- Gerson, S.L. MGMT: Its Role in Cancer Aetiology and Cancer Therapeutics. Nat. Rev. Cancer 2004, 4, 296–307. [Google Scholar] [CrossRef]
- Albertella, M.R.; Green, C.M.; Lehmann, A.R.; O’Connor, M.J. A Role for Polymerase Eta in the Cellular Tolerance to Cisplatin-Induced Damage. Cancer Res. 2005, 65, 9799–9806. [Google Scholar] [CrossRef] [Green Version]
- Doles, J.; Oliver, T.G.; Cameron, E.R.; Hsu, G.; Jacks, T.; Walker, G.C.; Hemann, M.T. Suppression of Rev3, the Catalytic Subunit of Polz, Sensitizes Drug-Resistant Lung Tumors to Chemotherapy. Proc. Natl. Acad. Sci. USA 2010, 107, 20786–20791. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Doles, J.; Hemann, M.T.; Walker, G.C. Error-Prone Translesion Synthesis Mediates Acquired Chemoresistance. Proc. Natl. Acad. Sci. USA 2010, 107, 20792–20797. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Zarth, A.T.; Carlson, E.S.; Villalta, P.W.; Upadhyaya, P.; Stepanov, I.; Hecht, S.S. Methyl DNA Phosphate Adduct Formation in Rats Treated Chronically with 4-(Methylnitrosamino)-1-(3-Pyridyl)-1-Butanone and Enantiomers of Its Metabolite 4-(Methylnitrosamino)-1-(3-Pyridyl)-1-Butanol. Chem. Res. Toxicol. 2018, 31, 48–57. [Google Scholar] [CrossRef]
- Jones, G.D.; Le Pla, R.C.; Farmer, P.B. Phosphotriester Adducts (PTEs): DNA’s Overlooked Lesion. Mutagenesis 2010, 25, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beranek, D.T. Distribution of Methyl and Ethyl Adducts Following Alkylation with Monofunctional Alkylating Agents. Mutat. Res. 1990, 231, 11–30. [Google Scholar] [PubMed]
- Ma, B.; Zarth, A.T.; Carlson, E.S.; Villalta, P.W.; Upadhyaya, P.; Stepanov, I.; Hecht, S.S. Identification of More than One Hundred Structurally Unique DNA-Phosphate Adducts Formed during Rat Lung Carcinogenesis by the Tobacco-Specific Nitrosamine 4-(Methylnitrosamino)-1-(3-Pyridyl)-1-Butanone. Carcinogenesis 2017, 39, 232–241. [Google Scholar] [CrossRef] [Green Version]
- Den Engelse, L.; De Graaf, A.; De Brij, R.J.; Menkveld, G.J. O2- and O4-Ethylthymine and the Ethylphosphotriester DTp(Et)DT Are Highly Persistent DNA Modifications in Slowly Dividing Tissues of the Ethylnitrosourea-Treated Rat. Carcinogenesis 1987, 8, 751–757. [Google Scholar] [CrossRef]
- Shooter, K.V.; Slade, T.A. The Stability of Methyl and Ethyl Phosphotriesters in DNA in Vivo. Chem. Biol. Interact. 1977, 19, 353–361. [Google Scholar] [CrossRef]
- Friedberg, E.C.; Aguilera, A.; Gellert, M.; Hanawalt, P.C.; Hays, J.B.; Lehmann, A.R.; Lindahl, T.; Lowndes, N.; Sarasin, A.; Wood, R.D. DNA Repair: From Molecular Mechanism to Human Disease. DNA Repair Amst. 2006, 5, 986–996. [Google Scholar] [CrossRef]
- Tan, Y.; Guo, S.; Wu, J.; Du, H.; Li, L.; You, C.; Wang, Y. DNA Polymerase η Promotes the Transcriptional Bypass of N2-Alkyl-2′-Deoxyguanosine Adducts in Human Cells. J. Am. Chem. Soc. 2021, 143, 16197–16205. [Google Scholar] [CrossRef]
- Wu, J.; Wang, P.; Wang, Y. Cytotoxic and Mutagenic Properties of Alkyl Phosphotriester Lesions in Escherichia Coli Cells. Nucleic Acids Res. 2018, 46, 4013–4021. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Li, L.; Wang, P.; You, C.; Williams, N.L.; Wang, Y. Translesion Synthesis of O4-Alkylthymidine Lesions in Human Cells. Nucleic Acids Res. 2016, 44, 9256–9265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Du, H.; Li, L.; Price, N.E.; Liu, X.; Wang, Y. The Impact of Minor-Groove N2-Alkyl-2′-Deoxyguanosine Lesions on DNA Replication in Human Cells. ACS Chem. Biol. 2019, 14, 1708–1716. [Google Scholar] [CrossRef]
- You, C.; Wang, Y. Quantitative Measurement of Transcriptional Inhibition and Mutagenesis Induced by Site-Specifically Incorporated DNA Lesions in Vitro and in Vivo. Nat. Protoc. 2015, 10, 1389–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; You, C.; Park, J.; Kim, H.S.; Guo, S.; Schärer, O.D.; Wang, Y. Transcriptional Perturbations of 2,6-Diaminopurine and 2-Aminopurine. ACS Chem. Biol. 2022, 17, 1672–1676. [Google Scholar] [CrossRef]
- Bregeon, D.; Doetsch, P.W. Transcriptional Mutagenesis: Causes and Involvement in Tumour Development. Nat. Rev. Cancer 2011, 11, 218–227. [Google Scholar] [CrossRef] [Green Version]
- Saxowsky, T.T.; Doetsch, P.W. RNA Polymerase Encounters with DNA Damage: Transcription-Coupled Repair or Transcriptional Mutagenesis? Chem. Rev. 2006, 106, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Morreall, J.F.; Petrova, L.; Doetsch, P.W. Transcriptional Mutagenesis and Its Potential Roles in the Etiology of Cancer and Bacterial Antibiotic Resistance. J. Cell. Physiol. 2013, 228, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Wang, Y. Replication of Pyridyloxobutyl Phosphotriester Lesions in Cells. Chem. Res. Toxicol. 2020, 33, 308–311. [Google Scholar] [CrossRef]
- Wu, J.; Yuan, J.; Price, N.E.; Wang, Y. Ada Protein- and Sequence Context-Dependent Mutagenesis of Alkyl Phosphotriester Lesions in Escherichia Coli Cells. J. Biol. Chem. 2020, 295, 8775–8783. [Google Scholar] [CrossRef] [PubMed]
- Koike, G.; Maki, H.; Takeya, H.; Hayakawa, H.; Sekiguchi, M. Purification, Structure, and Biochemical Properties of Human O6-Methylguanine-DNA Methyltransferase. J. Biol. Chem. 1990, 265, 14754–14762. [Google Scholar] [CrossRef]
- Viswanathan, A.; Doetsch, P.W. Effects of Nonbulky DNA Base Damages on Escherichia Coli RNA Polymerase-Mediated Elongation and Promoter Clearance. J. Biol. Chem. 1998, 273, 21276–21281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxowsky, T.T.; Meadows, K.L.; Klungland, A.; Doetsch, P.W. 8-Oxoguanine-Mediated Transcriptional Mutagenesis Causes Ras Activation in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18877–18882. [Google Scholar] [CrossRef] [Green Version]
- Lans, H.; Hoeijmakers, J.H.J.; Vermeulen, W.; Marteijn, J.A. The DNA Damage Response to Transcription Stress. Nat. Rev. Mol. Cell Biol. 2019, 20, 766–784. [Google Scholar] [CrossRef]
- Tornaletti, S.; Maeda, L.S.; Lloyd, D.R.; Reines, D.; Hanawalt, P.C. Effect of Thymine Glycol on Transcription Elongation by T7 RNA Polymerase and Mammalian RNA Polymerase II. J. Biol. Chem. 2001, 276, 45367–45371. [Google Scholar] [CrossRef]
- Charlet-Berguerand, N.; Feuerhahn, S.; Kong, S.E.; Ziserman, H.; Conaway, J.W.; Conaway, R.; Egly, J.M. RNA Polymerase II Bypass of Oxidative DNA Damage Is Regulated by Transcription Elongation Factors. EMBO J. 2006, 25, 5481–5491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clauson, C.L.; Oestreich, K.J.; Austin, J.W.; Doetsch, P.W. Abasic Sites and Strand Breaks in DNA Cause Transcriptional Mutagenesis in Escherichia Coli. Proc. Natl. Acad. Sci. USA 2010, 107, 3657–3662. [Google Scholar] [CrossRef] [Green Version]
- Tornaletti, S.; Reines, D.; Hanawalt, P.C. Structural Characterization of RNA Polymerase II Complexes Arrested by a Cyclobutane Pyrimidine Dimer in the Transcribed Strand of Template DNA. J. Biol. Chem. 1999, 274, 24124–24130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todd, R.C.; Lippard, S.J. Inhibition of Transcription by Platinum Antitumor Compounds. Metallomics 2009, 1, 280–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuasa, M.; Masutani, C.; Eki, T.; Hanaoka, F. Genomic Structure, Chromosomal Localization and Identification of Mutations in the Xeroderma Pigmentosum Variant (XPV) Gene. Oncogene 2000, 19, 4721–4728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biertümpfel, C.; Zhao, Y.; Kondo, Y.; Ramón-Maiques, S.; Gregory, M.; Lee, J.Y.; Masutani, C.; Lehmann, A.R.; Hanaoka, F.; Yang, W. Structure and Mechanism of Human DNA Polymerase η. Nature 2010, 465, 1044–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Egli, M.; Guengerich, F.P. Mechanism of Ribonucleotide Incorporation by Human DNA Polymerase η. J. Biol. Chem. 2016, 291, 3747–3756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Egli, M.; Guengerich, F.P. Human DNA Polymerase η Accommodates RNA for Strand Extension. J. Biol. Chem. 2017, 292, 18044–18051. [Google Scholar] [CrossRef] [Green Version]
- Mentegari, E.; Crespan, E.; Bavagnoli, L.; Kissova, M.; Bertoletti, F.; Sabbioneda, S.; Imhof, R.; Sturla, S.J.; Nilforoushan, A.; Hübscher, U.; et al. Ribonucleotide Incorporation by Human DNA Polymerase η Impacts Translesion Synthesis and RNase H2 Activity. Nucleic Acids Res. 2017, 45, 2600–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, A.; Nava, G.M.; Bianco, E.; Grasso, L.; Galati, E.; Bosio, M.C.; Delmastro, D.; Muzi-Falconi, M.; Lazzaro, F. RNase H Activities Counteract a Toxic Effect of Polymerase η in Cells Replicating with Depleted DNTP Pools. Nucleic Acids Res. 2019, 47, 4612–4623. [Google Scholar] [CrossRef]
- Gali, V.K.; Balint, E.; Serbyn, N.; Frittmann, O.; Stutz, F.; Unk, I. Translesion Synthesis DNA Polymerase η Exhibits a Specific RNA Extension Activity and a Transcription-Associated Function. Sci. Rep. 2017, 7, 13055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soria, G.; Belluscio, L.; van Cappellen, W.A.; Kanaar, R.; Essers, J.; Gottifredi, V. DNA Damage Induced Pol η Recruitment Takes Place Independently of the Cell Cycle Phase. Cell Cycle 2009, 8, 3340–3348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crespan, E.; Furrer, A.; Rösinger, M.; Bertoletti, F.; Mentegari, E.; Chiapparini, G.; Imhof, R.; Ziegler, N.; Sturla, S.J.; Hübscher, U.; et al. Impact of Ribonucleotide Incorporation by DNA Polymerases β and λ on Oxidative Base Excision Repair. Nat. Commun. 2016, 7, 10805. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Y.; Wu, J.; Clabaugh, G.; Li, L.; Du, H.; Wang, Y. Size- and Stereochemistry-Dependent Transcriptional Bypass of DNA Alkyl Phosphotriester Adducts in Mammalian Cells. DNA 2022, 2, 221-230. https://doi.org/10.3390/dna2040016
Tan Y, Wu J, Clabaugh G, Li L, Du H, Wang Y. Size- and Stereochemistry-Dependent Transcriptional Bypass of DNA Alkyl Phosphotriester Adducts in Mammalian Cells. DNA. 2022; 2(4):221-230. https://doi.org/10.3390/dna2040016
Chicago/Turabian StyleTan, Ying, Jiabin Wu, Garrit Clabaugh, Lin Li, Hua Du, and Yinsheng Wang. 2022. "Size- and Stereochemistry-Dependent Transcriptional Bypass of DNA Alkyl Phosphotriester Adducts in Mammalian Cells" DNA 2, no. 4: 221-230. https://doi.org/10.3390/dna2040016